Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs
- PMID: 32523949
- PMCID: PMC7261909
- DOI: 10.3389/fcell.2020.00347
Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs
Abstract
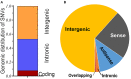
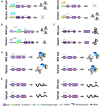
Single-nucleotide variants (SNVs) are the most common genetic variants and universally present in the human genome. Genome-wide association studies (GWASs) have identified a great number of disease or trait-associated variants, many of which are located in non-coding regions. Long intergenic non-protein coding RNAs (lincRNAs) are the major subtype of long non-coding RNAs; lincRNAs play crucial roles in various disorders and cellular models via multiple mechanisms. With rapid growth in the number of the identified lincRNAs and genetic variants, there is great demand for an investigation of SNVs in lincRNAs. Hence, in this article, we mainly summarize the significant role of SNVs within human lincRNA regions. Some pivotal variants may serve as risk factors for the development of various disorders, especially cancer. They may also act as important regulatory signatures involved in the modulation of lincRNAs in a tissue- or disorder-specific manner. An increasing number of researches indicate that lincRNA variants would potentially provide additional options for genetic testing and disease risk assessment in the personalized medicine era.
Keywords: biological function; disease susceptibility; long/large intergenic non-protein coding RNA; single-nucleotide variant; transcription.
Copyright © 2020 Zou, Wu, Tan, Shang and Zhou.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

