Genetic cluster analysis of SARS-CoV-2 and the identification of those responsible for the major outbreaks in various countries
- PMID: 32525765
- PMCID: PMC7477621
- DOI: 10.1080/22221751.2020.1773745
Genetic cluster analysis of SARS-CoV-2 and the identification of those responsible for the major outbreaks in various countries
Abstract
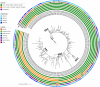
A newly emerged coronavirus, SARS-CoV-2, caused severe pneumonia outbreaks in China in December 2019 and has since spread to various countries around the world. To trace the evolution route and probe the transmission dynamics of this virus, we performed phylodynamic analysis of 247 high quality genomic sequences available in the GISAID platform as of 5 March 2020. Among them, four genetic clusters, defined as super-spreaders (SSs), could be identified and were found to be responsible for the major outbreaks that subsequently occurred in various countries. SS1 was widely disseminated in Asia and the US, and mainly responsible for outbreaks in the states of Washington and California as well as South Korea, whereas SS4 contributed to the pandemic in Europe. Using the signature mutations of each SS as markers, we further analysed 1539 genome sequences reported after 29 February 2020 and found that 90% of these genomes belonged to SSs, with SS4 being the most dominant. The relative degree of contribution of each SS to the pandemic in different continents was also depicted. Identification of these super-spreaders greatly facilitates development of new strategies to control the transmission of SARS-CoV-2.
Keywords: COVID-19; SARS-CoV-2; phylodynamic analysis; super-spreader; transmission.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Gorbalenya AE. Severe acute respiratory syndrome-related coronavirus – the species and its viruses, a statement of the Coronavirus Study Group. bioRxiv. 2020.
-
- Guan W-j, Ni Z-y, Hu Y, et al. . Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
