Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase
- PMID: 32526208
- PMCID: PMC7242921
- DOI: 10.1016/j.cell.2020.05.034
Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase
Abstract
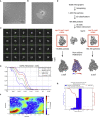
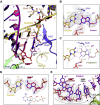
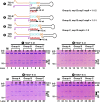
Nucleotide analog inhibitors, including broad-spectrum remdesivir and favipiravir, have shown promise in in vitro assays and some clinical studies for COVID-19 treatment, this despite an incomplete mechanistic understanding of the viral RNA-dependent RNA polymerase nsp12 drug interactions. Here, we examine the molecular basis of SARS-CoV-2 RNA replication by determining the cryo-EM structures of the stalled pre- and post- translocated polymerase complexes. Compared with the apo complex, the structures show notable structural rearrangements happening to nsp12 and its co-factors nsp7 and nsp8 to accommodate the nucleic acid, whereas there are highly conserved residues in nsp12, positioning the template and primer for an in-line attack on the incoming nucleotide. Furthermore, we investigate the inhibition mechanism of the triphosphate metabolite of remdesivir through structural and kinetic analyses. A transition model from the nsp7-nsp8 hexadecameric primase complex to the nsp12-nsp7-nsp8 polymerase complex is also proposed to provide clues for the understanding of the coronavirus transcription and replication machinery.
Keywords: 2019-nCoV; COVID-19; RdRP; SARS-CoV-2; favipiravir; nsp12; nsp8; polymerase; remdesivir; virus.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures












References
-
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018 doi: 10.1128/mBio.00221-18. - DOI - PMC - PubMed
-
- Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019 doi: 10.1128/JVI.01348-19. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

