Alterations in the Gut Microbiome and Suppression of Histone Deacetylases by Resveratrol Are Associated with Attenuation of Colonic Inflammation and Protection Against Colorectal Cancer
- PMID: 32526927
- PMCID: PMC7355848
- DOI: 10.3390/jcm9061796
Alterations in the Gut Microbiome and Suppression of Histone Deacetylases by Resveratrol Are Associated with Attenuation of Colonic Inflammation and Protection Against Colorectal Cancer
Abstract
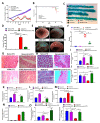
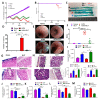
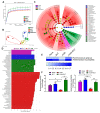
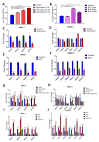
Inflammatory bowel disease (IBD) is known to significantly increase the risk for development of colorectal cancer (CRC), suggesting inflammation and cancer development are closely intertwined. Thus, agents that suppress inflammation may prevent the onset of cancer. In the current study, we used resveratrol, an anti-inflammatory stilbenoid, to study the role of microbiota in preventing inflammation-driven CRC. Resveratrol treatment in the azoxymethane (AOM) and dextran sodium sulphate (DSS) CRC murine model caused an increase in anti-inflammatory CD4 + FOXP3 + (Tregs) and CD4 + IL10 + cells, a decrease in proinflammatory Th1 and Th17 cells, and attenuated CRC development. Gut microbial profile studies demonstrated that resveratrol altered the gut microbiome and short chain fatty acid (SCFA), with modest increases in n-butyric acid and a potential butyrate precursor isobutyric acid. Fecal transfer from resveratrol-treated CRC mice and butyrate supplementation resulted in attenuation of disease and suppression of the inflammatory T cell response. Data also revealed both resveratrol and sodium butyrate (BUT) were capable of inhibiting histone deacetylases (HDACs), correlating with Treg induction. Analysis of The Cancer Genome Atlas (TCGA) datasets revealed increased expression of Treg-specific transcription factor FoxP3 or anti-inflammatory IL-10 resulted in an increase in 5-year survival of patients with CRC. These data suggest that alterations in the gut microbiome lead to an anti-inflammatory T cell response, leading to attenuation of inflammation-driven CRC.
Keywords: T regulatory cells; T-helper cells; azoxymethane; butyrate; colorectal cancer; dextran sodium sulfate; fecal transfer; histone deacetylase; microbiome; resveratrol.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Edwards B.K., Noone A.-M., Mariotto A.B., Simard E.P., Boscoe F.P., Henley S.J., Jemal A., Cho H., Anderson R.N., Kohler B.A., et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2013;120:1290–1314. doi: 10.1002/cncr.28509. - DOI - PMC - PubMed
-
- Bose D., Zimmerman L.J., Pierobon M., Petricoin E., Tozzi F., Parikh A., Fan F., Dallas N., Xia L., Gaur P., et al. Chemoresistant colorectal cancer cells and cancer stem cells mediate growth and survival of bystander cells. Br. J. Cancer. 2011;105:1759–1767. doi: 10.1038/bjc.2011.449. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

