Activity of Specialized Biomolecules against Gram-Positive and Gram-Negative Bacteria
- PMID: 32526972
- PMCID: PMC7344598
- DOI: 10.3390/antibiotics9060314
Activity of Specialized Biomolecules against Gram-Positive and Gram-Negative Bacteria
Abstract
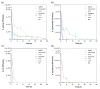
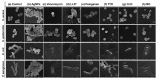
The increased resistance of bacteria against conventional pharmaceutical solutions, the antibiotics, has raised serious health concerns. This has stimulated interest in the development of bio-based therapeutics with limited resistance, namely, essential oils (EOs) or antimicrobial peptides (AMPs). This study envisaged the evaluation of the antimicrobial efficacy of selected biomolecules, namely LL37, pexiganan, tea tree oil (TTO), cinnamon leaf oil (CLO) and niaouli oil (NO), against four bacteria commonly associated to nosocomial infections: Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli and Pseudomonas aeruginosa. The antibiotic vancomycin and silver nanoparticles (AgNPs) were used as control compounds for comparison purposes. The biomolecules were initially screened for their antibacterial efficacy using the agar-diffusion test, followed by the determination of minimal inhibitory concentrations (MICs), kill-time kinetics and the evaluation of the cell morphology upon 24 h exposure. All agents were effective against the selected bacteria. Interestingly, the AgNPs required a higher concentration (4000-1250 µg/mL) to induce the same effects as the AMPs (500-7.8 µg/mL) or EOs (365.2-19.7 µg/mL). Pexiganan and CLO were the most effective biomolecules, requiring lower concentrations to kill both Gram-positive and Gram-negative bacteria (62.5-7.8 µg/mL and 39.3-19.7 µg/mL, respectively), within a short period of time (averaging 2 h 15 min for all bacteria). Most biomolecules apparently disrupted the bacteria membrane stability due to the observed cell morphology deformation and by effecting on the intracellular space. AMPs were observed to induce morphological deformations and cellular content release, while EOs were seen to split and completely envelope bacteria. Data unraveled more of the potential of these new biomolecules as replacements for the conventional antibiotics and allowed us to take a step forward in the understanding of their mechanisms of action against infection-related bacteria.
Keywords: antimicrobial peptides; bactericidal; essential oils; minimum inhibitory concentration; nosocomial.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Vapor-phase activities of cinnamon, thyme, and oregano essential oils and key constituents against foodborne microorganisms.J Agric Food Chem. 2007 May 30;55(11):4348-56. doi: 10.1021/jf063295u. Epub 2007 May 8. J Agric Food Chem. 2007. PMID: 17488023
-
Physical, Thermal, and Antibacterial Effects of Active Essential Oils with Potential for Biomedical Applications Loaded onto Cellulose Acetate/Polycaprolactone Wet-Spun Microfibers.Biomolecules. 2020 Jul 31;10(8):1129. doi: 10.3390/biom10081129. Biomolecules. 2020. PMID: 32751893 Free PMC article.
-
Coriander essential oil and linalool - interactions with antibiotics against Gram-positive and Gram-negative bacteria.Lett Appl Microbiol. 2019 Feb;68(2):156-164. doi: 10.1111/lam.13100. Epub 2019 Jan 4. Lett Appl Microbiol. 2019. PMID: 30471142
-
Sulopenem: An Intravenous and Oral Penem for the Treatment of Urinary Tract Infections Due to Multidrug-Resistant Bacteria.Drugs. 2022 Apr;82(5):533-557. doi: 10.1007/s40265-022-01688-1. Epub 2022 Mar 16. Drugs. 2022. PMID: 35294769 Review.
-
Antibacterial and antibiofilm potential of silver nanoparticles against antibiotic-sensitive and multidrug-resistant Pseudomonas aeruginosa strains.Braz J Microbiol. 2021 Mar;52(1):267-278. doi: 10.1007/s42770-020-00406-x. Epub 2020 Nov 24. Braz J Microbiol. 2021. PMID: 33231865 Free PMC article. Review.
Cited by
-
Synergistic nanocoating with layer-by-layer functionalized PCL membranes enhanced by manuka honey and essential oils for advanced wound healing.Sci Rep. 2024 Sep 5;14(1):20715. doi: 10.1038/s41598-024-71466-4. Sci Rep. 2024. PMID: 39237556 Free PMC article.
-
Effects of essential oils on economically important characteristics of ruminant species: A comprehensive review.Anim Nutr. 2023 Oct 20;16:1-10. doi: 10.1016/j.aninu.2023.05.017. eCollection 2024 Mar. Anim Nutr. 2023. PMID: 38131027 Free PMC article. Review.
-
Special Issue "Antimicrobial Biomaterials: Recent Progress".Int J Mol Sci. 2024 Jun 28;25(13):7153. doi: 10.3390/ijms25137153. Int J Mol Sci. 2024. PMID: 39000256 Free PMC article.
-
Comparison of Zinc Oxide Nanoparticle Integration into Non-Woven Fabrics Using Different Functionalisation Methods for Prospective Application as Active Facemasks.Polymers (Basel). 2023 Aug 22;15(17):3499. doi: 10.3390/polym15173499. Polymers (Basel). 2023. PMID: 37688127 Free PMC article.
-
The antimicrobial efficacy of rutin encapsulated chitosan versus multidrug-resistant Pseudomonas aeruginosa.Sci Rep. 2025 Jul 1;15(1):22047. doi: 10.1038/s41598-025-06873-2. Sci Rep. 2025. PMID: 40595077 Free PMC article.
References
-
- Quintiliani R.J., Sahm D., Courvalin P. Mechanisms of Resistance to Antimicrobial Agents. In: Murray P.R., Baron E.J., Pfaller M.A., Tenover F.C., Yolken R.H., editors. Manual of Clinical Microbiology. American Society for Microbiology; Washington, DC, USA: 1998. pp. 1505–1525.
-
- Mingeot-Leclercq M.-P., Décout J.-L. Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides. Med. Chem. Comm. 2016;7:586–611. doi: 10.1039/C5MD00503E. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

