Neurons that regulate mouse torpor
- PMID: 32528180
- PMCID: PMC7449701
- DOI: 10.1038/s41586-020-2387-5
Neurons that regulate mouse torpor
Abstract
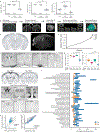
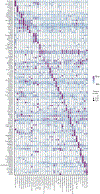
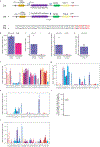
The advent of endothermy, which is achieved through the continuous homeostatic regulation of body temperature and metabolism1,2, is a defining feature of mammalian and avian evolution. However, when challenged by food deprivation or harsh environmental conditions, many mammalian species initiate adaptive energy-conserving survival strategies-including torpor and hibernation-during which their body temperature decreases far below its homeostatic set-point3-5. How homeothermic mammals initiate and regulate these hypothermic states remains largely unknown. Here we show that entry into mouse torpor, a fasting-induced state with a greatly decreased metabolic rate and a body temperature as low as 20 °C6, is regulated by neurons in the medial and lateral preoptic area of the hypothalamus. We show that restimulation of neurons that were activated during a previous bout of torpor is sufficient to initiate the key features of torpor, even in mice that are not calorically restricted. Among these neurons we identify a population of glutamatergic Adcyap1-positive cells, the activity of which accurately determines when mice naturally initiate and exit torpor, and the inhibition of which disrupts the natural process of torpor entry, maintenance and arousal. Taken together, our results reveal a specific neuronal population in the mouse hypothalamus that serves as a core regulator of torpor. This work forms a basis for the future exploration of mechanisms and circuitry that regulate extreme hypothermic and hypometabolic states, and enables genetic access to monitor, initiate, manipulate and study these ancient adaptations of homeotherm biology.
Figures

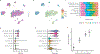








Comment in
-
Flipping the switch on the body's thermoregulatory system.Nature. 2020 Jul;583(7814):34-35. doi: 10.1038/d41586-020-01600-5. Nature. 2020. PMID: 32601488 No abstract available.
-
Chill out.Nat Rev Neurosci. 2020 Aug;21(8):397. doi: 10.1038/s41583-020-0337-8. Nat Rev Neurosci. 2020. PMID: 32612215 No abstract available.
References
-
- Morrison SF & Nakamura K. Central Mechanisms for Thermoregulation. Annu. Rev. Physiol 81, 285–308 (2019). - PubMed
-
- Heller HC & Hammel HT CNS control of body temperature during hibernation. Comp. Biochem. Physiol. A Physiol. 41, 349–359 (1972). - PubMed
-
- Geiser F. Metabolic Rate and Body Temperature Reduction During Hibernation and Daily Torpor. Annu. Rev. Physiol 66, 239–274 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

