Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome
- PMID: 32528815
- PMCID: PMC7282418
- DOI: 10.24171/j.phrp.2020.11.3.05
Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome
Abstract
Objectives: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, in December 2019 and has been rapidly spreading worldwide. Although the causal relationship among mutations and the features of SARS-CoV-2 such as rapid transmission, pathogenicity, and tropism, remains unclear, our results of genomic mutations in SARS-CoV-2 may help to interpret the interaction between genomic characterization in SARS-CoV-2 and infectivity with the host.
Methods: A total of 4,254 genomic sequences of SARS-CoV-2 were collected from the Global Initiative on Sharing all Influenza Data (GISAID). Multiple sequence alignment for phylogenetic analysis and comparative genomic approach for mutation analysis were conducted using Molecular Evolutionary Genetics Analysis (MEGA), and an in-house program based on Perl language, respectively.
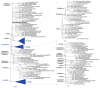
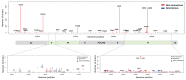
Results: Phylogenetic analysis of SARS-CoV-2 strains indicated that there were 3 major clades including S, V, and G, and 2 subclades (G.1 and G.2). There were 767 types of synonymous and 1,352 types of non-synonymous mutation. ORF1a, ORF1b, S, and N genes were detected at high frequency, whereas ORF7b and E genes exhibited low frequency. In the receptor-binding domain (RBD) of the S gene, 11 non-synonymous mutations were observed in the region adjacent to the angiotensin converting enzyme 2 (ACE2) binding site.
Conclusion: It has been reported that the rapid infectivity and transmission of SARS-CoV-2 associated with host receptor affinity are derived from several mutations in its genes. Without these genetic mutations to enhance evolutionary adaptation, species recognition, host receptor affinity, and pathogenicity, it would not survive. It is expected that our results could provide an important clue in understanding the genomic characteristics of SARS-CoV-2.
Keywords: SARS-CoV-2; evolutions; mutation.
Copyright ©2020, Korea Centers for Disease Control and Prevention.
Conflict of interest statement
Conflicts of Interest The authors have no conflicts of interest to declare.
Figures






References
-
- The Johns Hopkins Center for Health Security. nCoV Genetics [Internet] [cited 2020 Feb 3]. Available from: http://www.centerforhealthsecurity.org/resources/COVID-19/COVID-19-fact-....
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

