Serological differentiation between COVID-19 and SARS infections
- PMID: 32529906
- PMCID: PMC7473126
- DOI: 10.1080/22221751.2020.1780951
Serological differentiation between COVID-19 and SARS infections
Abstract
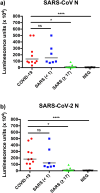
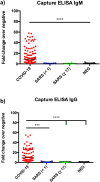
In response to the coronavirus disease 2019 (COVID-19) outbreak, caused by SARS-CoV-2, multiple diagnostic tests are required for acute disease diagnosis, contact tracing, monitoring asymptomatic infection rates and assessing herd immunity. While PCR remains the frontline test of choice in the acute diagnostic setting, serological tests are urgently needed. Unlike PCR tests which are highly specific, cross-reactivity is a major challenge for COVID-19 antibody tests considering there are six other coronaviruses known to infect humans. SARS-CoV is genetically related to SARS-CoV-2 sharing approximately 80% sequence identity and both belong to the species SARS related coronavirus in the genus Betacoronavirus of family Coronaviridae. We developed and compared the performance of four different serological tests to comprehensively assess the cross-reactivity between COVID-19 and SARS patient sera. There is significant cross-reactivity when N protein of either virus is used. The S1 or RBD regions from the spike (S) protein offers better specificity. Amongst the different platforms, capture ELISA performed best. We found that SARS survivors all have significant levels of antibodies remaining in their blood 17 years after infection. Anti-N antibodies waned more than anti-RBD antibodies, and the latter is known to play a more important role in providing protective immunity.
Keywords: COVID-19; SARS; SARS-CoV-2; antibody; serology.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. WHO, COVID-19 status report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio...., 2020. Accessed on 1 June 2020. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
