Tuning Transcription Factor Availability through Acetylation-Mediated Genomic Redistribution
- PMID: 32531202
- PMCID: PMC7427332
- DOI: 10.1016/j.molcel.2020.05.025
Tuning Transcription Factor Availability through Acetylation-Mediated Genomic Redistribution
Abstract
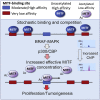
It is widely assumed that decreasing transcription factor DNA-binding affinity reduces transcription initiation by diminishing occupancy of sequence-specific regulatory elements. However, in vivo transcription factors find their binding sites while confronted with a large excess of low-affinity degenerate motifs. Here, using the melanoma lineage survival oncogene MITF as a model, we show that low-affinity binding sites act as a competitive reservoir in vivo from which transcription factors are released by mitogen-activated protein kinase (MAPK)-stimulated acetylation to promote increased occupancy of their regulatory elements. Consequently, a low-DNA-binding-affinity acetylation-mimetic MITF mutation supports melanocyte development and drives tumorigenesis, whereas a high-affinity non-acetylatable mutant does not. The results reveal a paradoxical acetylation-mediated molecular clutch that tunes transcription factor availability via genome-wide redistribution and couples BRAF to tumorigenesis. Our results further suggest that p300/CREB-binding protein-mediated transcription factor acetylation may represent a common mechanism to control transcription factor availability.
Keywords: DNA-binding affinity; E-box; MITF; acetylation; bHLH-LZ; melanocyte; melanoma; transcription factor.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Bertolotto C., Lesueur F., Giuliano S., Strub T., de Lichy M., Bille K., Dessen P., d’Hayer B., Mohamdi H., Remenieras A., French Familial Melanoma Study Group A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. - PubMed
-
- Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. - PubMed
-
- Blackwell T.K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
-
- Boyes J., Byfield P., Nakatani Y., Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

