Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype
- PMID: 32531372
- PMCID: PMC7283064
- DOI: 10.1016/j.jaci.2020.05.051
Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype
Abstract
Background: More than 300 million people carry a diagnosis of asthma, with data to suggest that they are at a higher risk for infection or adverse outcomes from severe acute respiratory syndrome coronavirus 2. Asthma is remarkably heterogeneous, and it is currently unclear how patient-intrinsic factors may relate to coronavirus disease 2019.
Objective: We sought to identify and characterize subsets of patients with asthma at increased risk for severe acute respiratory syndrome coronavirus 2 infection.
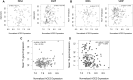
Methods: Participants from 2 large asthma cohorts were stratified using clinically relevant parameters to identify factors related to angiotensin-converting enzyme-2 (ACE2) expression within bronchial epithelium. ACE-2-correlated gene signatures were used to interrogate publicly available databases to identify upstream signaling events and novel therapeutic targets.
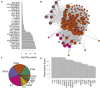
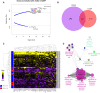
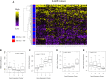
Results: Stratifying by type 2 inflammatory biomarkers, we identified subjects who demonstrated low peripheral blood eosinophils accompanied by increased expression of the severe acute respiratory syndrome coronavirus 2 receptor ACE2 in bronchial epithelium. Genes highly correlated with ACE2 overlapped with type 1 and 2 IFN signatures, normally induced by viral infections. T-cell recruitment and activation within bronchoalveolar lavage cells of ACE2-high subjects was reciprocally increased. These patients demonstrated characteristics corresponding to risk factors for severe coronavirus disease 2019, including male sex, history of hypertension, low peripheral blood, and elevated bronchoalveolar lavage lymphocytes.
Conclusions: ACE2 expression is linked to upregulation of viral response genes in a subset of type 2-low patients with asthma with characteristics resembling known risk factors for severe coronavirus disease 2019. Therapies targeting the IFN family and T-cell-activating factors may therefore be of benefit in a subset of patients.
Keywords: ACE2; COVID-19; SARS-CoV-2; Type-2 low; asthma; coronavirus; interferons; viral response.
Copyright © 2020 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan [April 12, 2020]. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.04.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

