Linking Biochemical and Structural States of SERCA: Achievements, Challenges, and New Opportunities
- PMID: 32532023
- PMCID: PMC7313052
- DOI: 10.3390/ijms21114146
Linking Biochemical and Structural States of SERCA: Achievements, Challenges, and New Opportunities
Abstract
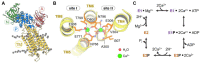
Sarcoendoplasmic reticulum calcium ATPase (SERCA), a member of the P-type ATPase family of ion and lipid pumps, is responsible for the active transport of Ca2+ from the cytoplasm into the sarcoplasmic reticulum lumen of muscle cells, into the endoplasmic reticulum (ER) of non-muscle cells. X-ray crystallography has proven to be an invaluable tool in understanding the structural changes of SERCA, and more than 70 SERCA crystal structures representing major biochemical states (defined by bound ligand) have been deposited in the Protein Data Bank. Consequently, SERCA is one of the best characterized components of the calcium transport machinery in the cell. Emerging approaches in the field, including spectroscopy and molecular simulation, now help integrate and interpret this rich structural information to understand the conformational transitions of SERCA that occur during activation, inhibition, and regulation. In this review, we provide an overview of the crystal structures of SERCA, focusing on identifying metrics that facilitate structure-based categorization of major steps along the catalytic cycle. We examine the integration of crystallographic data with different biophysical approaches and computational methods to link biochemical and structural states of SERCA that are populated in the cell. Finally, we discuss the challenges and new opportunities in the field, including structural elucidation of functionally important and novel regulatory complexes of SERCA, understanding the structural basis of functional divergence among homologous SERCA regulators, and bridging the gap between basic and translational research directed toward therapeutic modulation of SERCA.
Keywords: P-type ATPase; SERCA; X-ray crystallography; biophysical methods; calcium; inhibition; molecular simulation; regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

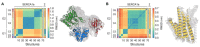







Similar articles
-
Structural basis for relief of phospholamban-mediated inhibition of the sarcoplasmic reticulum Ca2+-ATPase at saturating Ca2+ conditions.J Biol Chem. 2018 Aug 10;293(32):12405-12414. doi: 10.1074/jbc.RA118.003752. Epub 2018 Jun 22. J Biol Chem. 2018. PMID: 29934304 Free PMC article.
-
The SarcoEndoplasmic Reticulum Calcium ATPase.Subcell Biochem. 2018;87:229-258. doi: 10.1007/978-981-10-7757-9_8. Subcell Biochem. 2018. PMID: 29464562 Review.
-
Atomic-level characterization of the activation mechanism of SERCA by calcium.PLoS One. 2011;6(10):e26936. doi: 10.1371/journal.pone.0026936. Epub 2011 Oct 27. PLoS One. 2011. PMID: 22046418 Free PMC article.
-
Conformational Transitions and Alternating-Access Mechanism in the Sarcoplasmic Reticulum Calcium Pump.J Mol Biol. 2017 Mar 10;429(5):647-666. doi: 10.1016/j.jmb.2017.01.007. Epub 2017 Jan 16. J Mol Biol. 2017. PMID: 28093226 Free PMC article.
-
A diversity of SERCA Ca2+ pump inhibitors.Biochem Soc Trans. 2011 Jun;39(3):789-97. doi: 10.1042/BST0390789. Biochem Soc Trans. 2011. PMID: 21599650 Review.
Cited by
-
Protein Adsorption on Solid Supported Membranes: Monitoring the Transport Activity of P-Type ATPases.Molecules. 2020 Sep 11;25(18):4167. doi: 10.3390/molecules25184167. Molecules. 2020. PMID: 32933017 Free PMC article. Review.
-
A Computational Swiss Army Knife Approach to Unraveling the Secrets of Proton Movement through SERCA.Biophys J. 2020 Sep 1;119(5):890-891. doi: 10.1016/j.bpj.2020.07.028. Epub 2020 Aug 6. Biophys J. 2020. PMID: 32795397 Free PMC article. No abstract available.
-
Targeting oncogenic Notch signaling with SERCA inhibitors.J Hematol Oncol. 2021 Jan 6;14(1):8. doi: 10.1186/s13045-020-01015-9. J Hematol Oncol. 2021. PMID: 33407740 Free PMC article. Review.
-
Mutational analysis of an antimalarial drug target, PfATP4.Proc Natl Acad Sci U S A. 2025 Jan 14;122(2):e2403689122. doi: 10.1073/pnas.2403689122. Epub 2025 Jan 8. Proc Natl Acad Sci U S A. 2025. PMID: 39773028 Free PMC article.
-
Machine Learning-Driven Discovery of Structurally Related Natural Products as Activators of the Cardiac Calcium Pump SERCA2a.ChemMedChem. 2025 May 5;20(9):e202400913. doi: 10.1002/cmdc.202400913. Epub 2025 Feb 6. ChemMedChem. 2025. PMID: 39853697 Free PMC article.
References
-
- Gorski P.A., Ceholski D.K., Young H.S. Membrane Dynamics and Calcium Signaling. Springer International Publishing; Berlin/Heidelberg, Germany: 2017. Structure-Function Relationship of the SERCA Pump and Its Regulation by Phospholamban and Sarcolipin; pp. 77–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

