Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs-A Resistant Animal of Prion Disease
- PMID: 32532135
- PMCID: PMC7311962
- DOI: 10.3390/ijms21114160
Novel Polymorphisms and Genetic Characteristics of the Prion Protein Gene (PRNP) in Dogs-A Resistant Animal of Prion Disease
Abstract
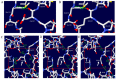
Transmissible spongiform encephalopathies (TSEs) have been reported in a wide range of species. However, TSE infection in natural cases has never been reported in dogs. Previous studies have reported that polymorphisms of the prion protein gene (PRNP) have a direct impact on the susceptibility of TSE. However, studies on polymorphisms of the canine PRNP gene are very rare in dogs. We examined the genotype, allele, and haplotype frequencies of canine PRNP in 204 dogs using direct sequencing and analyzed linkage disequilibrium (LD) using Haploview version 4.2. In addition, to evaluate the impact of nonsynonymous polymorphisms on the function of prion protein (PrP), we carried out in silico analysis using PolyPhen-2, PROVEAN, and PANTHER. Furthermore, we analyzed the structure of PrP and hydrogen bonds according to alleles of nonsynonymous single nucleotide polymorphisms (SNPs) using the Swiss-Pdb Viewer program. Finally, we predicted the impact of the polymorphisms on the aggregation propensity of dog PrP using AMYCO. We identified a total of eight polymorphisms, including five novel SNPs and one insertion/deletion polymorphism, and found strong LDs and six major haplotypes among eight polymorphisms. In addition, we identified significantly different distribution of haplotypes among eight dog breeds, however, the kinds of identified polymorphisms were different among each dog breed. We predicted that p.64_71del HGGGWGQP, Asp182Gly, and Asp182Glu polymorphisms can impact the function and/or structure of dog PrP. Furthermore, the number of hydrogen bonds of dog PrP with the Glu182 and Gly182 alleles were predicted to be less than those with the Asp182 allele. Finally, Asp163Glu and Asp182Gly showed more aggregation propensity than wild-type dog PrP. These results suggest that nonsynonymous SNPs, Asp182Glu and Asp182Gly, can influence the stability of dog PrP and confer the possibility of TSE infection in dogs.
Keywords: PRNP; SNP; dog; octapeptide; polymorphism; prion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Novel Polymorphisms and Genetic Features of the Prion Protein Gene (PRNP) in Cats, Hosts of Feline Spongiform Encephalopathy.Genes (Basel). 2020 Dec 24;12(1):13. doi: 10.3390/genes12010013. Genes (Basel). 2020. PMID: 33374431 Free PMC article.
-
Absence of Strong Genetic Linkage Disequilibrium between Single Nucleotide Polymorphisms (SNPs) in the Prion Protein Gene (PRNP) and the Prion-Like Protein Gene (PRND) in the Horse, a Prion-Resistant Species.Genes (Basel). 2020 May 7;11(5):518. doi: 10.3390/genes11050518. Genes (Basel). 2020. PMID: 32392732 Free PMC article.
-
Identification of the novel polymorphisms and potential genetic features of the prion protein gene (PRNP) in horses, a prion disease-resistant animal.Sci Rep. 2020 Jun 2;10(1):8926. doi: 10.1038/s41598-020-65731-5. Sci Rep. 2020. PMID: 32488112 Free PMC article.
-
Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies.Dongwuxue Yanjiu. 2014 Sep;35(5):436-45. doi: 10.13918/j.issn.2095-8137.2014.5.436. Dongwuxue Yanjiu. 2014. PMID: 25297084 Free PMC article. Review.
-
Association of the PRNP regulatory region polymorphisms with the occurrence of sporadic Creutzfeldt-Jakob disease.Folia Neuropathol. 2012;50(1):68-73. Folia Neuropathol. 2012. PMID: 22505365 Review.
Cited by
-
Assessing Different Chronic Wasting Disease Training Aids for Use with Detection Dogs.Animals (Basel). 2024 Jan 18;14(2):300. doi: 10.3390/ani14020300. Animals (Basel). 2024. PMID: 38254469 Free PMC article.
-
Identification of a novel risk factor for chronic wasting disease (CWD) in elk: S100G single nucleotide polymorphism (SNP) of the prion protein gene (PRNP).Vet Res. 2023 Jun 16;54(1):48. doi: 10.1186/s13567-023-01177-7. Vet Res. 2023. PMID: 37328789 Free PMC article.
-
First report of a novel polymorphism and genetic characteristics of the leporine prion protein (PRNP) gene.Front Vet Sci. 2023 Sep 22;10:1229369. doi: 10.3389/fvets.2023.1229369. eCollection 2023. Front Vet Sci. 2023. PMID: 37808111 Free PMC article.
-
First report of structural characteristics and polymorphisms of the prion protein gene in raccoon dogs: The possibility of prion disease-resistance.Front Vet Sci. 2022 Sep 20;9:989352. doi: 10.3389/fvets.2022.989352. eCollection 2022. Front Vet Sci. 2022. PMID: 36204297 Free PMC article.
-
Identification of the Highly Polymorphic Prion Protein Gene (PRNP) in Frogs (Rana dybowskii).Animals (Basel). 2025 Jan 15;15(2):220. doi: 10.3390/ani15020220. Animals (Basel). 2025. PMID: 39858220 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

