Quantification of ongoing APOBEC3A activity in tumor cells by monitoring RNA editing at hotspots
- PMID: 32532990
- PMCID: PMC7293259
- DOI: 10.1038/s41467-020-16802-8
Quantification of ongoing APOBEC3A activity in tumor cells by monitoring RNA editing at hotspots
Abstract
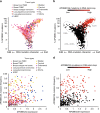
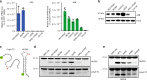
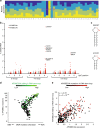
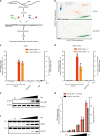
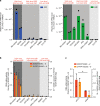
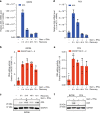
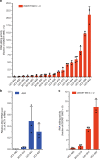
APOBEC3A is a cytidine deaminase driving mutagenesis, DNA replication stress and DNA damage in cancer cells. While the APOBEC3A-induced vulnerability of cancers offers an opportunity for therapy, APOBEC3A protein and mRNA are difficult to quantify in tumors due to their low abundance. Here, we describe a quantitative and sensitive assay to measure the ongoing activity of APOBEC3A in tumors. Using hotspot RNA mutations identified from APOBEC3A-positive tumors and droplet digital PCR, we develop an assay to quantify the RNA-editing activity of APOBEC3A. This assay is superior to APOBEC3A protein- and mRNA-based assays in predicting the activity of APOBEC3A on DNA. Importantly, we demonstrate that the RNA mutation-based APOBEC3A assay is applicable to clinical samples from cancer patients. Our study presents a strategy to follow the dysregulation of APOBEC3A in tumors, providing opportunities to investigate the role of APOBEC3A in tumor evolution and to target the APOBEC3A-induced vulnerability in therapy.
Conflict of interest statement
L.Z. has received research funding from Calico. R.B.C. is a consultant/advisory board member for Amgen, Array Biopharma, Astex Pharmaceuticals, Avidity Biosciences, BMS, C4 Therapeutics, Chugai, Elicio, Fog Pharma, Fount Therapeutics, Genentech, Guardant Health, LOXO, Merrimack, N-of-one, Novartis, nRichDx, Revolution Medicines, Roche, Roivant, Shionogi, Shire, Spectrum Pharmaceuticals, Symphogen, Taiho, and Warp Drive Bio; holds equity in Avidity Biosciences, C4 Therapeutics, Fount Therapeutics, nRichDx, and Revolution Medicines; and has received research funding from Asana, AstraZeneca, and Sanofi. P.J., D.B., A.L., S.P., K.A., A.G.F., M.S.L., and R.B. declare no competing interest.
Figures
References
-
- Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

