Epigenetic and epitranscriptomic regulation of viral replication
- PMID: 32533130
- PMCID: PMC7291935
- DOI: 10.1038/s41579-020-0382-3
Epigenetic and epitranscriptomic regulation of viral replication
Abstract
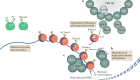
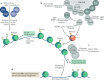
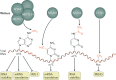
Eukaryotic gene expression is regulated not only by genomic enhancers and promoters, but also by covalent modifications added to both chromatin and RNAs. Whereas cellular gene expression may be either enhanced or inhibited by specific epigenetic modifications deposited on histones (in particular, histone H3), these epigenetic modifications can also repress viral gene expression, potentially functioning as a potent antiviral innate immune response in DNA virus-infected cells. However, viruses have evolved countermeasures that prevent the epigenetic silencing of their genes during lytic replication, and they can also take advantage of epigenetic silencing to establish latent infections. By contrast, the various covalent modifications added to RNAs, termed epitranscriptomic modifications, can positively regulate mRNA translation and/or stability, and both DNA and RNA viruses have evolved to utilize epitranscriptomic modifications as a means to maximize viral gene expression. As a consequence, both chromatin and RNA modifications could serve as novel targets for the development of antivirals. In this Review, we discuss how host epigenetic and epitranscriptomic processes regulate viral gene expression at the levels of chromatin and RNA function, respectively, and explore how viruses modify, avoid or utilize these processes in order to regulate viral gene expression.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

