Biofilm dispersion
- PMID: 32533131
- PMCID: PMC8564779
- DOI: 10.1038/s41579-020-0385-0
Biofilm dispersion
Abstract
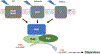
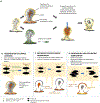
The formation of microbial biofilms enables single planktonic cells to assume a multicellular mode of growth. During dispersion, the final step of the biofilm life cycle, single cells egress from the biofilm to resume a planktonic lifestyle. As the planktonic state is considered to be more vulnerable to antimicrobial agents and immune responses, dispersion is being considered a promising avenue for biofilm control. In this Review, we discuss conditions that lead to dispersion and the mechanisms by which native and environmental cues contribute to dispersion. We also explore recent findings on the role of matrix degradation in the dispersion process, and the distinct phenotype of dispersed cells. Last, we discuss the translational and therapeutic potential of dispersing bacteria during infection.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

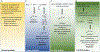
References
-
- Costerton JW, Stewart PS & Greenberg EP Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999). - PubMed
-
- Geesey GG, Richardson WT, Yeomans HG, Irvin RT & Costerton JW Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol 23, 1733–1736 (1977). - PubMed
-
- Costerton JW et al. Bacterial biofilms in nature and disease. Annual Reviews in Microbiology 41, 435–464 (1987). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

