Noninvasive Fetal Genotyping by Droplet Digital PCR to Identify Maternally Inherited Monogenic Diabetes Variants
- PMID: 32533152
- PMCID: PMC7611030
- DOI: 10.1093/clinchem/hvaa104
Noninvasive Fetal Genotyping by Droplet Digital PCR to Identify Maternally Inherited Monogenic Diabetes Variants
Abstract
Background: Babies of women with heterozygous pathogenic glucokinase (GCK) variants causing mild fasting hyperglycemia are at risk of macrosomia if they do not inherit the variant. Conversely, babies who inherit a pathogenic hepatocyte nuclear factor 4α (HNF4A) diabetes variant are at increased risk of high birth weight. Noninvasive fetal genotyping for maternal pathogenic variants would inform pregnancy management.
Methods: Droplet digital PCR was used to quantify reference and variant alleles in cell-free DNA extracted from blood from 38 pregnant women heterozygous for a GCK or HNF4A variant and to determine fetal fraction by measurement of informative maternal and paternal variants. Droplet numbers positive for the reference/alternate allele together with the fetal fraction were used in a Bayesian analysis to derive probability for the fetal genotype. The babies' genotypes were ascertained postnatally by Sanger sequencing.
Results: Droplet digital PCR assays for GCK or HNF4A variants were validated for testing in all 38 pregnancies. Fetal fraction of ≥2% was demonstrated in at least 1 cell-free DNA sample from 33 pregnancies. A threshold of ≥0.95 for calling homozygous reference genotypes and ≤0.05 for heterozygous fetal genotypes allowed correct genotype calls for all 33 pregnancies with no false-positive results. In 30 of 33 pregnancies, a result was obtained from a single blood sample.
Conclusions: This assay can be used to identify pregnancies at risk of macrosomia due to maternal monogenic diabetes variants.
Keywords: Bayesian analysis; droplet digital PCR; monogenic diabetes; non-invasive prenatal diagnosis.
© American Association for Clinical Chemistry 2020.
Conflict of interest statement
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/orpotential conflicts of interest:
Figures
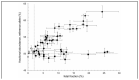

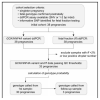
References
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence offetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. - PubMed
-
- Boon EM, Faas BH. Benefits and limitations of whole genome versus targeted approaches for noninvasive prenatal testing for fetal aneuploidies. Prenat Diagn. 2013;33:563–8. - PubMed
-
- Hyett JA, Gardener G, Stojilkovic-Mikic T, Finning KM, Martin PG, Rodeck CH, Chitty LS. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat Diagn. 2005;25:1111–6. - PubMed
-
- Clausen FB. Integration of noninvasive prenatal prediction of fetal blood group into clinical prenatal care. Prenat Diagn. 2014;34:409–15. - PubMed
-
- Chitty LS, Griffin DR, Meaney C, Barrett A, Khalil A, Pajkrt E, Cole TJ. New aids for the non-invasive prenatal diagnosis of achondroplasia: dysmorphic features, charts of fetal size and molecular confirmation using cell-free fetal DNA in maternal plasma. Ultrasound Obstet Gynecol. 2011;37:283–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

