Dynamics of Ligand Binding to a Rigid Glycosidase*
- PMID: 32533782
- PMCID: PMC7693232
- DOI: 10.1002/anie.202003236
Dynamics of Ligand Binding to a Rigid Glycosidase*
Abstract
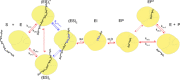
The single-domain GH11 glycosidase from Bacillus circulans (BCX) is involved in the degradation of hemicellulose, which is one of the most abundant renewable biomaterials in nature. We demonstrate that BCX in solution undergoes minimal structural changes during turnover. NMR spectroscopy results show that the rigid protein matrix provides a frame for fast substrate binding in multiple conformations, accompanied by slow conversion, which is attributed to an enzyme-induced substrate distortion. A model is proposed in which the rigid enzyme takes advantage of substrate flexibility to induce a conformation that facilitates the acyl formation step of the hydrolysis reaction.
Keywords: NMR spectroscopy; dynamics; glycosidases; ligand binding; rigid fold.
© 2020 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

