In The Blood: Connecting Variant to Function In Human Hematopoiesis
- PMID: 32534791
- PMCID: PMC7363574
- DOI: 10.1016/j.tig.2020.05.006
In The Blood: Connecting Variant to Function In Human Hematopoiesis
Abstract
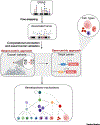
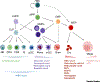
Genome-wide association studies (GWAS) have identified thousands of genetic variants associated with a range of human diseases and traits. However, understanding the mechanisms by which these genetic variants have an impact on associated diseases and traits, often referred to as the variant-to-function (V2F) problem, remains a significant hurdle. Solving the V2F challenge requires us to identify causative genetic variants, relevant cell types/states, target genes, and mechanisms by which variants can cause diseases or alter phenotypic traits. We discuss emerging functional approaches that are being applied to tackle the V2F problem for blood cell traits, illuminating how human genetic variation can impact on key mechanisms in hematopoiesis, as well as highlighting future prospects for this nascent field.
Keywords: GWAS; blood cell traits; hematopoiesis; variant to function.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures

References
-
- Sankaran VG et al. (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322 (5909), 1839–42. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

