Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study
- PMID: 32536605
- PMCID: PMC7873428
- DOI: 10.1136/gutjnl-2020-321106
Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study
Abstract
Objective: The microbiome contributes to the pathogenesis of inflammatory bowel disease (IBD) but the relative contribution of different lifestyle and environmental factors to the compositional variability of the gut microbiota is unclear.
Design: Here, we rank the size effect of disease activity, medications, diet and geographic location of the faecal microbiota composition (16S rRNA gene sequencing) in patients with Crohn's disease (CD; n=303), ulcerative colitis (UC; n = 228) and controls (n=161), followed longitudinally (at three time points with 16 weeks intervals).
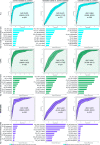
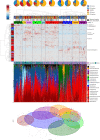
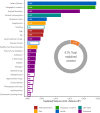
Results: Reduced microbiota diversity but increased variability was confirmed in CD and UC compared with controls. Significant compositional differences between diseases, particularly CD, and controls were evident. Longitudinal analyses revealed reduced temporal microbiota stability in IBD, particularly in patients with changes in disease activity. Machine learning separated disease from controls, and active from inactive disease, when consecutive time points were modelled. Geographic location accounted for most of the microbiota variance, second to the presence or absence of CD, followed by history of surgical resection, alcohol consumption and UC diagnosis, medications and diet with most (90.3%) of the compositional variance stochastic or unexplained.
Conclusion: The popular concept of precision medicine and rational design of any therapeutic manipulation of the microbiota will have to contend not only with the heterogeneity of the host response, but also with widely differing lifestyles and with much variance still unaccounted for.
Keywords: Crohn's disease; colonic microflora; diet; ulcerative colitis.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: MTB reports grants and personal fees from AbbVie Canada, grants and personal fees from Janssen Canada, grants and personal fees from Pfizer Canada, grants from Shire Canada, grants and personal fees from Takeda Canada, personal fees from Mylan Pharmaceuticals, other from AbbVie, Janssen, Pfizer, Boerhinger Ingelheim, Celgene, outside the submitted work; FS reports other from Alimentary Health/Precision Biotics, other from 4D pharma, Cork, other from Atlantia Food Clinical Trials, personal fees from Kaleido Biosciences, outside the submitted work; MJC reports personal fees from Mars PetCare, other from Second Genome, outside the submitted work.
Figures




Comment in
-
Overcoming regional limitations: transfer learning for cross-regional microbial-based diagnosis of diseases.Gut. 2023 Oct;72(10):2004-2006. doi: 10.1136/gutjnl-2022-328216. Epub 2022 Oct 28. Gut. 2023. PMID: 36307182 Free PMC article. No abstract available.
-
Microbiome variance of the small bowel in Crohn's disease.Gut. 2023 Aug;72(8):1626-1628. doi: 10.1136/gutjnl-2022-327313. Epub 2022 Sep 1. Gut. 2023. PMID: 37414433 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
