Comparison of cattle BoLA-DRB3 typing by PCR-RFLP, direct sequencing, and high-resolution DNA melting curve analysis
- PMID: 32537103
- PMCID: PMC7282214
- DOI: 10.30466/vrf.2018.90444.2189
Comparison of cattle BoLA-DRB3 typing by PCR-RFLP, direct sequencing, and high-resolution DNA melting curve analysis
Abstract
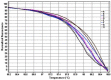
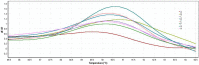
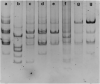
Major histocompatibility complex (MHC) represents an important genetic marker for manipulation to improve the health and productivity of cattle. It is closely associated with numerous disease susceptibilities and immune responses. Bovine MHC, also called bovine leukocyte antigen (BoLA), is considered as a suitable marker for genetic diversity studies. In cattle, most of the polymorphisms are located in exon 2 of BoLA-DRB3, which encodes the peptide-binding cleft. In this study, the polymorphism of the BoLA-DRB3.2 gene in Holstein's calves was studied using high resolution melting curve analysis (HRM). Observed HRM results were compared to PCR-RFLP and direct sequencing techniques. Eight different HRM and seven different RFLP profiles were identified among the population studied. By comparing to sequencing data, HRM could completely discriminate all genotypes (eight profiles), while the RFLP failed to distinguish between the genotypes *1101/*1001 and *1104/*1501. According to the results, the HRM analysis method gave more accurate results than RFLP by differentiating between the BoLA-DRB3.2 genotypes. Due to the Co-dominant nature of the MHC alleles, HRM technique could be used for investigating the polymorphisms of genotypes and their associations with immune responses.
Keywords: BoLA-DRB3.2; Genotyping; HRM; Holstein; RFLP.
© 2020 Urmia University. All rights reserved.
Figures
References
-
- Gorer PA. The genetic and antigenic basis of tumour transplantation. J Pathol. 1937;44(3):691–697.
-
- Bonner J. Major histocompatibility complex influences reproductive efficiency: Evolutionary implications. J Craniofac Genet Dev Biol Suppl. 1986;2:5–11. - PubMed
-
- Penn DJ, Potts WK. The evolution of mating preferences and major histocompatibility complex genes. Am Nat. 1999;153(2):145–164. - PubMed
-
- Graham RR, Ortmann W, Rodine P, et al. Specific combinations of HLA-DR2 and DR3 class II haplotypes contribute graded risk for disease susceptibility and autoantibodies in human SLE. Eur J Hum Genet. 2007;15(8):823–830. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials