Glutaredoxin AtGRXS8 represses transcriptional and developmental responses to nitrate in Arabidopsis thaliana roots
- PMID: 32537558
- PMCID: PMC7287413
- DOI: 10.1002/pld3.227
Glutaredoxin AtGRXS8 represses transcriptional and developmental responses to nitrate in Arabidopsis thaliana roots
Abstract
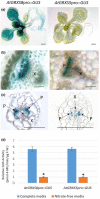
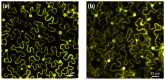
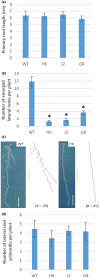
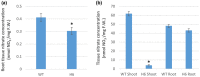
Glutaredoxins (GRXs) are small oxidoreductase enzymes that can reduce disulfide bonds in target proteins. The class III GRX gene family is unique to land plants, and Arabidopsis thaliana has 21 class III GRXs, which remain largely uncharacterized. About 80% of A. thaliana class III GRXs are transcriptionally regulated by nitrate, and several recent studies have suggested roles for these GRXs in nitrogen signaling. Our objective was to functionally characterize two nitrate-induced GRX genes, AtGRXS5 and AtGRXS8, defining their roles in signaling and development in the A. thaliana root. We demonstrated that AtGRXS5 and AtGRXS8 are primarily expressed in root and shoot vasculature (phloem), and that the corresponding GRX proteins display nucleo-cytosolic subcellular localization. Ectopic expression of AtGRXS8 in transgenic plants caused major alterations in root system architecture: Normal primary root development, but a near absence of lateral roots. RNA sequencing demonstrated that the roots of AtGRXS8-overexpressing plants show strongly reduced transcript abundance for many primary nitrate response genes, including the major high-affinity nitrate transporters. Correspondingly, high-affinity nitrate uptake and the transport of nitrate from roots to shoots are compromised in AtGRXS8-overexpressing plants. Finally, we demonstrated that the AtGRXS8 protein can physically interact with the TGA1 and TGA4 transcription factors, which are central regulators of early transcriptional responses to nitrate in A. thaliana roots. Overall, these results suggest that AtGRXS8 acts to quench both transcriptional and developmental aspects of primary nitrate response, potentially by interfering with the activity of the TGA1 and TGA4 transcription factors.
Keywords: Arabidopsis thaliana; glutaredoxin; nitrate; root development; signaling.
© 2020 The Authors. Plant Direct published by American Society of Plant Biologists, Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest associated with the work described in this manuscript.
Figures



References
-
- Alvarez, J. M. , Riveras, E. , Vidal, E. A. Gras, D. E. , Contreras‐López, O. , Tamayo, K. P. , … Jordana, X. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal, 80, 1–13. - PubMed
-
- Bae, M. S. , Cho, E. J. , Choi, E.‐Y. , & Park, O. K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. The Plant Journal, 36, 652–663. - PubMed
-
- Bartel, P. L. , & Fields, S. (1995) [16] Analyzing protein‐protein interactions using two‐hybrid system In Methods in Enzymology. Oncogene techniques (pp. 241–263). Academic Press: Retrieved from http://www.sciencedirect.com/science/article/pii/0076687995540180 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

