Structural Enzymology of Nitrogenase Enzymes
- PMID: 32538623
- PMCID: PMC8606229
- DOI: 10.1021/acs.chemrev.0c00067
Structural Enzymology of Nitrogenase Enzymes
Abstract
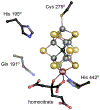
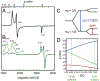
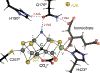
The reduction of dinitrogen to ammonia by nitrogenase reflects a complex choreography involving two component proteins, MgATP and reductant. At center stage of this process resides the active site cofactor, a complex metallocluster organized around a trigonal prismatic arrangement of iron sites surrounding an interstitial carbon. As a consequence of the choreography, electrons and protons are delivered to the active site for transfer to the bound N2. While the detailed mechanism for the substrate reduction remains enigmatic, recent developments highlight the role of hydrides and the privileged role for two irons of the trigonal prism in the binding of exogenous ligands. Outstanding questions concern the precise nature of the intermediates between N2 and NH3, and whether the cofactor undergoes significant rearrangement during turnover; resolution of these issues will require the convergence of biochemistry, structure, spectroscopy, computation, and model chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








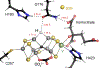
References
-
- Guth JH; Burris RH Inhibition of Nitrogenase-Catalyzed NH3 Formation by H2. Biochemistry 1983, 22 (22), 5111. - PubMed
-
- Snider MG; Temple BS; Wolfenden R The path to the transition state in enzyme reactions: a survey of catalytic efficiencies. J. Phys. Org. Chem. 2004, 17 (6–7), 586.
-
- Eady RR Structure-function relationships of alternative nitrogenases. Chem. Rev. 1996, 96 (7), 3013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

