Porcine circovirus type 2 (PCV2) genotyping in Austrian pigs in the years 2002 to 2017
- PMID: 32539835
- PMCID: PMC7294622
- DOI: 10.1186/s12917-020-02413-4
Porcine circovirus type 2 (PCV2) genotyping in Austrian pigs in the years 2002 to 2017
Abstract
Background: Eight different PCV2 genotypes with varying prevalence and clinical impact have been described so far. PCV2 infection is still widespread among the vaccinated population and several experimental studies have clearly demonstrated that there is no induction of a 100% cross-protective immunity between the PCV2 genotypes. Hence, PCV2a-based vaccines may be ineffective. In this longitudinal study, the PCV2 genotype and haplotype evolution in Austria in the years 2002 to 2017 was investigated by phylogenetic analysis of 462 bp-long sequences of the capsid protein gene (ORF2). The obtained findings may be of practical relevance for the future development of vaccination strategies.
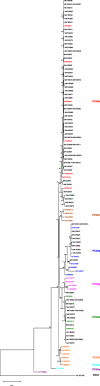
Results: One hundred thirty four of a total of 161 formalin-fixed and paraffin wax-embedded samples could be sequenced successfully. There was no significant influence of storage time on sequencing success or quality. PCV2a (8.2%), PCV2b (77.6%), PCV2d (13.4%), and PCV2g (0.8%) were found. PCV2d was first detected as early as in 2004. PCV2g was described once in 2009. Both global PCV2 genotype shifts were observed. PCV2a occurred with a low prevalence during the first study years only in samples from non-vaccinated swine herds and was gradually replaced by PCV2b until 2011. PCV2b was the most prevalent genotype over the whole study period and was detected in samples from vaccinated and non-vaccinated herds. During the last two study years, the prevalence of PCV2d increased, although at this point almost all herds were vaccinated. The haplotype diversity was high, but the nucleotide diversity was low. Especially for genotype PCV2b, an increase in haplotype diversity could be described during the first study years.
Conclusion: Extensive PCV2a-derived vaccination resulted in a reduction of prevalence and in a stabilization of genotype PCV2a, whereas genotypes PCV2b and PCV2d evolved as a consequence of natural and vaccination-induced selection. An ongoing virus circulation may be the result of reduced vaccine-induced protection.
Keywords: Genotype shift; Genotypes; Haplotypes; Porcine circovirus type 2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Allan GM, McNeilly F, Kennedy S, Daft B, Clark EG, Ellis JA, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Investig. 1998;10(1):3–10. - PubMed
-
- Xiao CT, Halbur PG, Opriessnig T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol. 2015;96(7):1830–1841. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

