Eltrombopag is a potential target for drug intervention in SARS-CoV-2 spike protein
- PMID: 32540428
- PMCID: PMC7290210
- DOI: 10.1016/j.meegid.2020.104419
Eltrombopag is a potential target for drug intervention in SARS-CoV-2 spike protein
Abstract
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is a current global threat for which there is an urgent need to search for an effective therapy. The transmembrane spike (S) glycoprotein of SARS-CoV-2 directly binds to the host angiotensin-converting enzyme 2 (ACE2) and mediates viral entrance, which is therefore considered as a promising drug target. Considering that new drug development is a time-consuming process, drug repositioning may facilitate rapid drug discovery dealing with sudden infectious diseases. Here, we compared the differences between the virtual structural proteins of SARS-CoV-2 and SARS-CoV, and selected a pocket mainly localizing in the fusion cores of S2 domain for drug screening. A virtual drug design algorithm screened the Food and Drug Administration-approved drug library of 1234 compounds, and 13 top scored compounds were obtained through manual screening. Through in vitro molecular interaction experiments, eltrombopag was further verified to possess a high binding affinity to S protein plus human ACE2 and could potentially affect the stability of the ACE2-S protein complex. Hence, it is worth further exploring eltrombopag as a potential drug for the treatment of SARS-CoV-2 infection.
Keywords: Homology Modeling; SARS-CoV-2; Surface plasmon resonance; Virtual drug design.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
None.
Figures


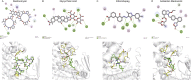





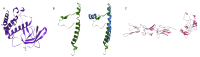


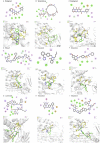


References
-
- Bonora E., et al. Studies on the mechanism of action of sulfonylureas in type-ii diabetic subjects - gliquidone. J. Endocrinol. Investig. 1992;15(1):1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

