The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain
- PMID: 32540970
- PMCID: PMC7397115
- DOI: 10.1074/jbc.RA120.013976
The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain
Abstract
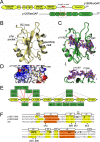
The Src homology 2 (SH2) domain has a highly conserved architecture that recognizes linear phosphotyrosine motifs and is present in a wide range of signaling pathways across different evolutionary taxa. A hallmark of SH2 domains is the arginine residue in the conserved FLVR motif that forms a direct salt bridge with bound phosphotyrosine. Here, we solve the X-ray crystal structures of the C-terminal SH2 domain of p120RasGAP (RASA1) in its apo and peptide-bound form. We find that the arginine residue in the FLVR motif does not directly contact pTyr1087 of a bound phosphopeptide derived from p190RhoGAP; rather, it makes an intramolecular salt bridge to an aspartic acid. Unexpectedly, coordination of phosphotyrosine is achieved by a modified binding pocket that appears early in evolution. Using isothermal titration calorimetry, we find that substitution of the FLVR arginine R377A does not cause a significant loss of phosphopeptide binding, but rather a tandem substitution of R398A (SH2 position βD4) and K400A (SH2 position βD6) is required to disrupt the binding. These results indicate a hitherto unrecognized diversity in SH2 domain interactions with phosphotyrosine and classify the C-terminal SH2 domain of p120RasGAP as "FLVR-unique."
Keywords: FLVR motif; GTPase-activating protein (GAP); RASA1; RasGAP; Src homology 2 domain (SH2 domain); X-ray crystallography; p120RasGAP; phosphotyrosine; protein structure; protein–protein interaction.
© 2020 Jaber Chehayeb et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures
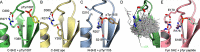

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

