EYES ABSENT and TIMELESS integrate photoperiodic and temperature cues to regulate seasonal physiology in Drosophila
- PMID: 32541062
- PMCID: PMC7334534
- DOI: 10.1073/pnas.2004262117
EYES ABSENT and TIMELESS integrate photoperiodic and temperature cues to regulate seasonal physiology in Drosophila
Abstract
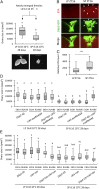
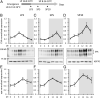
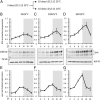
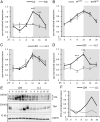
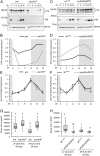
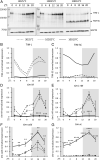
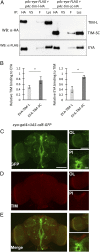
Organisms possess photoperiodic timing mechanisms to detect variations in day length and temperature as the seasons progress. The nature of the molecular mechanisms interpreting and signaling these environmental changes to elicit downstream neuroendocrine and physiological responses are just starting to emerge. Here, we demonstrate that, in Drosophila melanogaster, EYES ABSENT (EYA) acts as a seasonal sensor by interpreting photoperiodic and temperature changes to trigger appropriate physiological responses. We observed that tissue-specific genetic manipulation of eya expression is sufficient to disrupt the ability of flies to sense seasonal cues, thereby altering the extent of female reproductive dormancy. Specifically, we observed that EYA proteins, which peak at night in short photoperiod and accumulate at higher levels in the cold, promote reproductive dormancy in female D. melanogaster Furthermore, we provide evidence indicating that the role of EYA in photoperiodism and temperature sensing is aided by the stabilizing action of the light-sensitive circadian clock protein TIMELESS (TIM). We postulate that increased stability and level of TIM at night under short photoperiod together with the production of cold-induced and light-insensitive TIM isoforms facilitate EYA accumulation in winter conditions. This is supported by our observations that tim null mutants exhibit reduced incidence of reproductive dormancy in simulated winter conditions, while flies overexpressing tim show an increased incidence of reproductive dormancy even in long photoperiod.
Keywords: alternative splicing; circadian clock; photoperiod; seasonality; temperature.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Emerson K. J., Bradshaw W. E., Holzapfel C. M., Complications of complexity: Integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 25, 217–225 (2009). - PubMed
-
- Bonini N. M., Leiserson W. M., Benzer S., The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379–395 (1993). - PubMed
-
- Duncan M. K. et al. ., Eyes absent: A gene family found in several metazoan phyla. Mamm. Genome 8, 479–485 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

