Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly
- PMID: 32541867
- PMCID: PMC7608415
- DOI: 10.1038/s41422-020-0353-2
Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly
Erratum in
-
Author Corrections: Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly.Cell Res. 2021 Dec;31(12):1319-1320. doi: 10.1038/s41422-021-00550-5. Cell Res. 2021. PMID: 34381185 Free PMC article. No abstract available.
Abstract
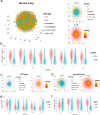
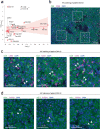
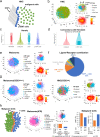
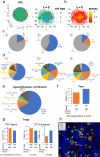
Single-cell RNA sequencing (scRNA-seq) has revolutionized transcriptomic studies by providing unprecedented cellular and molecular throughputs, but spatial information of individual cells is lost during tissue dissociation. While imaging-based technologies such as in situ sequencing show great promise, technical difficulties currently limit their wide usage. Here we hypothesize that cellular spatial organization is inherently encoded by cell identity and can be reconstructed, at least in part, by ligand-receptor interactions, and we present CSOmap, a computational tool to infer cellular interaction de novo from scRNA-seq. We show that CSOmap can successfully recapitulate the spatial organization of multiple organs of human and mouse including tumor microenvironments for multiple cancers in pseudo-space, and reveal molecular determinants of cellular interactions. Further, CSOmap readily simulates perturbation of genes or cell types to gain novel biological insights, especially into how immune cells interact in the tumor microenvironment. CSOmap can be a widely applicable tool to interrogate cellular organizations based on scRNA-seq data for various tissues in diverse systems.
Conflict of interest statement
Z.Z. is a founder of Analytical Biosciences Limited. Other authors declare no competing interests.
Figures




References
-
- Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proc. Soc. Exp. Biol. Med. 1941;47:200–202. doi: 10.3181/00379727-47-13084P. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

