FOXO1 promotes HIV latency by suppressing ER stress in T cells
- PMID: 32541947
- PMCID: PMC7483895
- DOI: 10.1038/s41564-020-0742-9
FOXO1 promotes HIV latency by suppressing ER stress in T cells
Abstract
Quiescence is a hallmark of CD4+ T cells latently infected with human immunodeficiency virus 1 (HIV-1). While reversing this quiescence is an effective approach to reactivate latent HIV from T cells in culture, it can cause deleterious cytokine dysregulation in patients. As a key regulator of T-cell quiescence, FOXO1 promotes latency and suppresses productive HIV infection. We report that, in resting T cells, FOXO1 inhibition impaired autophagy and induced endoplasmic reticulum (ER) stress, thereby activating two associated transcription factors: activating transcription factor 4 (ATF4) and nuclear factor of activated T cells (NFAT). Both factors associate with HIV chromatin and are necessary for HIV reactivation. Indeed, inhibition of protein kinase R-like ER kinase, an ER stress sensor that can mediate the induction of ATF4, and calcineurin, a calcium-dependent regulator of NFAT, synergistically suppressed HIV reactivation induced by FOXO1 inhibition. Thus, our studies uncover a link of FOXO1, ER stress and HIV infection that could be therapeutically exploited to selectively reverse T-cell quiescence and reduce the size of the latent viral reservoir.
Conflict of interest statement
Competing interests
The authors declare that there is no conflict of interest.
Figures




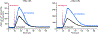




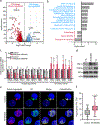
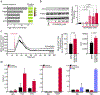

References
-
- Barré-Sinoussi F, Ross AL & Delfraissy J-F Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol 11, 877–883 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

