Transcriptome sequencing reveals signatures of positive selection in the Spot-Tailed Earless Lizard
- PMID: 32542006
- PMCID: PMC7295237
- DOI: 10.1371/journal.pone.0234504
Transcriptome sequencing reveals signatures of positive selection in the Spot-Tailed Earless Lizard
Abstract
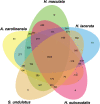
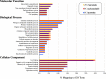
The continual loss of threatened biodiversity is occurring at an accelerated pace. High-throughput sequencing technologies are now providing opportunities to address this issue by aiding in the generation of molecular data for many understudied species of high conservation interest. Our overall goal of this study was to begin building the genomic resources to continue investigations and conservation of the Spot-Tailed Earless lizard. Here we leverage the power of high-throughput sequencing to generate the liver transcriptome for the Northern Spot-Tailed Earless Lizard (Holbrookia lacerata) and Southern Spot-Tailed Earless Lizard (Holbrookia subcaudalis), which have declined in abundance in the past decades, and their sister species, the Common Lesser Earless Lizard (Holbrookia maculata). Our efforts produced high quality and robust transcriptome assemblies validated by 1) quantifying the number of processed reads represented in the transcriptome assembly and 2) quantifying the number of highly conserved single-copy orthologs that are present in our transcript set using the BUSCO pipeline. We found 1,361 1-to-1 orthologs among the three Holbrookia species, Anolis carolinensis, and Sceloporus undulatus. We carried out dN/dS selection tests using a branch-sites model and identified a dozen genes that experienced positive selection in the Holbrookia lineage with functions in development, immunity, and metabolism. Our single-copy orthologous sequences additionally revealed significant pairwise sequence divergence (~.73%) between the Northern H. lacerata and Southern H. subcaudalis that further supports the recent elevation of the Southern Spot-Tailed Earless Lizard to full species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Phylogenetic structure of Holbrookia lacerata (Cope 1880) (Squamata: Phrynosomatidae): one species or two?Zootaxa. 2019 Jun 18;4619(1):zootaxa.4619.1.6. doi: 10.11646/zootaxa.4619.1.6. Zootaxa. 2019. PMID: 31716318
-
The Coachella valley fringe-toed lizard (Uma inornata): genetic diversity and phylogenetic relationships of an endangered species.Mol Phylogenet Evol. 2001 Mar;18(3):327-34. doi: 10.1006/mpev.2000.0881. Mol Phylogenet Evol. 2001. PMID: 11277627
-
METASTATIC MINERALIZATION IN A ZOOLOGIC COLLECTION OF SPOT-TAILED EARLESS LIZARDS (HOLBROOKIASPP.).J Zoo Wildl Med. 2023 Mar;54(1):175-184. doi: 10.1638/2021-0173. J Zoo Wildl Med. 2023. PMID: 36971643
-
Phylogenetic relationships and heterogeneous evolutionary processes among phrynosomatine sand lizards (Squamata, Iguanidae) revisited.Mol Phylogenet Evol. 2008 May;47(2):700-16. doi: 10.1016/j.ympev.2008.01.010. Epub 2008 Jan 24. Mol Phylogenet Evol. 2008. PMID: 18362078
-
Genome-guided transcriptome assembly in the age of next-generation sequencing.IEEE/ACM Trans Comput Biol Bioinform. 2013 Sep-Oct;10(5):1234-40. doi: 10.1109/tcbb.2013.140. IEEE/ACM Trans Comput Biol Bioinform. 2013. PMID: 24524156 Free PMC article. Review.
Cited by
-
Insights into the species evolution of Calanus copepods in the northern seas revealed by de novo transcriptome sequencing.Ecol Evol. 2022 Feb 22;12(2):e8606. doi: 10.1002/ece3.8606. eCollection 2022 Feb. Ecol Evol. 2022. PMID: 35228861 Free PMC article.
References
-
- Roelke CE, Maldonado JA, Pope BW, Firneno TJ, Laduc TJ, Hibbitts TJ, et al. Mitochondrial genetic variation within and between Holbrookia lacerata lacerata and Holbrookia lacerata subcaudalis, the spot-tailed earless lizards of Texas. J Nat Hist. 2018;52: 1017–1027.
-
- Frankham R, Briscoe DA, Ballou JD. Introduction to Conservation Genetics. Cambridge University Press; 2002.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

