Frequency and spectrum of disease-causing variants in 1892 patients with suspected genetic HLH disorders
- PMID: 32542393
- PMCID: PMC7322966
- DOI: 10.1182/bloodadvances.2020001605
Frequency and spectrum of disease-causing variants in 1892 patients with suspected genetic HLH disorders
Abstract
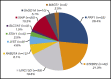
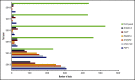
This article explores the distribution and mutation spectrum of potential disease-causing genetic variants in hemophagocytic lymphohistiocytosis (HLH)-associated genes observed in a large tertiary clinical referral laboratory. Samples from 1892 patients submitted for HLH genetic analysis were studied between September 2013 and June 2018 using a targeted next-generation sequencing panel approach. Patients ranged in age from 1 day to 78 years. Analysis included 15 genes associated with HLH. A potentially causal genetic finding was observed in 227 (12.0%) samples in this cohort. A total of 197 patients (10.4%) had a definite genetic diagnosis. Patients with pathogenic variants in familial HLH genes tended to be diagnosed significantly younger compared with other genes. Pathogenic or likely pathogenic variants in the PRF1 gene were the most frequent. However, mutations in genes associated with degranulation defects (STXBP2, UNC13D, RAB27A, LYST, and STX11) were more common than previously appreciated and collectively represented >50% of cases. X-linked conditions (XIAP, SH2D1A, and MAGT1) accounted for 17.8% of the 197 cases. Pathogenic variants in the SLC7A7 gene were the least encountered. These results describe the largest cohort of genetic variation associated with suspected HLH in North America. Merely 10.4% of patients were identified with a clearly genetic cause by this diagnostic approach; other possible etiologies of HLH should be investigated. These results suggest that careful thought should be given regarding whether patients have a clinical phenotype most consistent with HLH vs other clinical and disease phenotypes. The gene panel identified known pathogenic and novel variants in 10 HLH-associated genes.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
-
- Stepp SE, Dufourcq-Lagelouse R, Le Deist F, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286(5446):1957-1959. - PubMed
-
- Feldmann J, Callebaut I, Raposo G, et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3). Cell. 2003;115(4):461-473. - PubMed
-
- zur Stadt U, Schmidt S, Kasper B, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14(6):827-834. - PubMed

