Several ways one goal-methanogenesis from unconventional substrates
- PMID: 32542472
- PMCID: PMC7374477
- DOI: 10.1007/s00253-020-10724-7
Several ways one goal-methanogenesis from unconventional substrates
Abstract
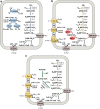
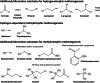
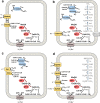
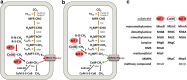
Methane is the second most important greenhouse gas on earth. It is produced by methanogenic archaea, which play an important role in the global carbon cycle. Three main methanogenesis pathways are known: in the hydrogenotrophic pathway H2 and carbon dioxide are used for methane production, whereas in the methylotrophic pathway small methylated carbon compounds like methanol and methylated amines are used. In the aceticlastic pathway, acetate is disproportionated to methane and carbon dioxide. However, next to these conventional substrates, further methanogenic substrates and pathways have been discovered. Several phylogenetically distinct methanogenic lineages (Methanosphaera, Methanimicrococcus, Methanomassiliicoccus, Methanonatronarchaeum) have evolved hydrogen-dependent methylotrophic methanogenesis without the ability to perform either hydrogenotrophic or methylotrophic methanogenesis. Genome analysis of the deep branching Methanonatronarchaeum revealed an interesting membrane-bound hydrogenase complex affiliated with the hardly described class 4 g of multisubunit hydrogenases possibly providing reducing equivalents for anabolism. Furthermore, methylated sulfur compounds such as methanethiol, dimethyl sulfide, and methylmercaptopropionate were described to be converted into adapted methylotrophic methanogenesis pathways of Methanosarcinales strains. Moreover, recently it has been shown that the methanogen Methermicoccus shengliensis can use methoxylated aromatic compounds in methanogenesis. Also, tertiary amines like choline (N,N,N-trimethylethanolamine) or betaine (N,N,N-trimethylglycine) have been described as substrates for methane production in Methanococcoides and Methanolobus strains. This review article will provide in-depth information on genome-guided metabolic reconstructions, physiology, and biochemistry of these unusual methanogenesis pathways. KEY POINTS: • Newly discovered methanogenic substrates and pathways are reviewed for the first time. • The review provides an in-depth analysis of unusual methanogenesis pathways. • The hydrogenase complex of the deep branching Methanonatronarchaeum is analyzed.
Keywords: Archaea; Extended substrate range; Methane production; Novel pathways.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Asakawa S, Sauer K, Liesack W, Tauer RK. Tetramethylammonium:coenzyme M methyltransferase system from Methanococcoides sp. Arch Microbiol. 1998;170:220–226. - PubMed
-
- Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261.
-
- Bak F, Finster K, Rothfuss F. Formation of dimethylsulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol. 1992;157:529–534.
-
- Bond DR, Lovley DR. Reduction of Fe(III) oxide by methanogens in the presence and absence of extracellular quinones. Environ Microbiol. 2002;4:115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

