Loss of NF2 defines a genetic subgroup of non-FOS-rearranged osteoblastoma
- PMID: 32542935
- PMCID: PMC7578308
- DOI: 10.1002/cjp2.172
Loss of NF2 defines a genetic subgroup of non-FOS-rearranged osteoblastoma
Abstract
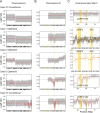
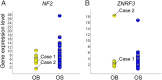
Osteoblastoma is a locally aggressive tumour of bone. Until recently, its underlying genetic features were largely unknown. During the past two years, reports have demonstrated that acquired structural variations affect the transcription factor FOS in a high proportion of cases. These rearrangements modify the terminal exon of the gene and are believed to stabilise both the FOS transcript and the encoded protein, resulting in high expression levels. Here, we applied in-depth genetic analyses to a series of 29 osteoblastomas, including five classified as epithelioid osteoblastoma. We found recurrent homozygous deletions of the NF2 gene in three of the five epithelioid cases and in one conventional osteoblastoma. These events were mutually exclusive from FOS mutations. Structural variations were determined by deep whole genome sequencing and the number of FOS-rearranged cases was less than previously reported (10/23, 43%). One conventional osteoblastoma displayed a novel mechanism of FOS upregulation; bringing the entire FOS gene under the control of the WNT5A enhancer that is itself activated by FOS. Taken together, we show that NF2 loss characterises a subgroup of osteoblastomas, distinct from FOS-rearranged cases. Both NF2 and FOS are involved in regulating bone homeostasis, thereby providing a mechanistic link to the excessive bone growth of osteoblastoma.
Keywords: FOS; FOSB; NF2; WNT5A; osteoblastoma; osteosarcoma.
© 2020 The Authors. The Journal of Pathology: Clinical Research published by The Pathological Society of Great Britain and Ireland & John Wiley & Sons, Ltd.
Figures

References
-
- Amary F, Markert E, Berisha F, et al FOS expression in osteoid osteoma and osteoblastoma: a valuable ancillary diagnostic tool. Am J Surg Pathol 2019; 43: 1661–1667. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

