PKD2/polycystin-2 induces autophagy by forming a complex with BECN1
- PMID: 32543276
- PMCID: PMC8354594
- DOI: 10.1080/15548627.2020.1782035
PKD2/polycystin-2 induces autophagy by forming a complex with BECN1
Abstract
Macroautophagy/autophagy is an intracellular process involved in the breakdown of macromolecules and organelles. Recent studies have shown that PKD2/PC2/TRPP2 (polycystin 2, transient receptor potential cation channel), a nonselective cation channel permeable to Ca2+ that belongs to the family of transient receptor potential channels, is required for autophagy in multiple cell types by a mechanism that remains unclear. Here, we report that PKD2 forms a protein complex with BECN1 (beclin 1), a key protein required for the formation of autophagic vacuoles, by acting as a scaffold that interacts with several co-modulators via its coiled-coil domain (CCD). Our data identified a physical and functional interaction between PKD2 and BECN1, which depends on one out of two CCD domains (CC1), located in the carboxy-terminal tail of PKD2. In addition, depletion of intracellular Ca2+ with BAPTA-AM not only blunted starvation-induced autophagy but also disrupted the PKD2-BECN1 complex. Consistently, PKD2 overexpression triggered autophagy by increasing its interaction with BECN1, while overexpression of PKD2D509V, a Ca2+ channel activity-deficient mutant, did not induce autophagy and manifested diminished interaction with BECN1. Our findings show that the PKD2-BECN1 complex is required for the induction of autophagy, and its formation depends on the presence of the CC1 domain of PKD2 and on intracellular Ca2+ mobilization by PKD2. These results provide new insights regarding the molecular mechanisms by which PKD2 controls autophagy.Abbreviations: ADPKD: autosomal dominant polycystic kidney disease; ATG: autophagy-related; ATG14/ATG14L: autophagy related 14; Baf A1: bafilomycin A1; BCL2/Bcl-2: BCL2 apoptosis regulator; BCL2L1/BCL-XL: BCL2 like 1; BECN1: beclin 1; CCD: coiled-coil domain; EBSS: Earle's balanced salt solution; ER: endoplasmic reticulum; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GFP: green fluorescent protein; GOLGA2/GM130: golgin A2; GST: glutathione s-transferase; LAMP1: lysosomal associated membrane protein 1; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MTORC1: mechanistic target of rapamycin kinase complex 1; NBR1: NBR1 autophagy cargo receptor; PIK3C3/VPS34: phosphatidylinositol 3-kinase catalytic subunit type 3; PKD2/PC2: polycystin 2, transient receptor potential cation channel; RTN4/NOGO: reticulon 4; RUBCN/RUBICON: rubicon autophagy regulator; SQSTM1/p62: sequestosome 1; UVRAG: UV radiation resistance associated; WIPI2: WD repeat domain, phosphoinositide interacting 2.
Keywords: Autophagy; beclin 1; calcium; polycystin-2; protein complex.
Conflict of interest statement
No potential conflict of interest was reported by the authors
Figures

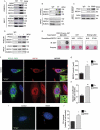
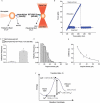
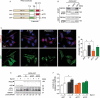
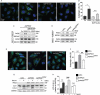
References
-
- Onodera J, Ohsumi Y.. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280(36):31582–31586. - PubMed
-
- Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5(7):455–468. - PubMed
-
- Klionsky DJ, Cregg JM, Dunn WA, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
