Bacterial chromosome segregation by the ParABS system
- PMID: 32543349
- PMCID: PMC7333895
- DOI: 10.1098/rsob.200097
Bacterial chromosome segregation by the ParABS system
Abstract
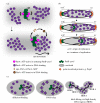
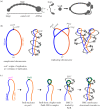
Proper chromosome segregation during cell division is essential in all domains of life. In the majority of bacterial species, faithful chromosome segregation is mediated by the tripartite ParABS system, consisting of an ATPase protein ParA, a CTPase and DNA-binding protein ParB, and a centromere-like parS site. The parS site is most often located near the origin of replication and is segregated first after chromosome replication. ParB nucleates on parS before binding to adjacent non-specific DNA to form a multimeric nucleoprotein complex. ParA interacts with ParB to drive the higher-order ParB-DNA complex, and hence the replicating chromosomes, to each daughter cell. Here, we review the various models for the formation of the ParABS complex and describe its role in segregating the origin-proximal region of the chromosome. Additionally, we discuss outstanding questions and challenges in understanding bacterial chromosome segregation.
Keywords: ParA–ParB–parS; SMC; chromosome maintenance; chromosome organization; chromosome segregation; spreading.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
