3D model of harlequin ichthyosis reveals inflammatory therapeutic targets
- PMID: 32544098
- PMCID: PMC7456239
- DOI: 10.1172/JCI132987
3D model of harlequin ichthyosis reveals inflammatory therapeutic targets
Abstract
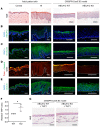
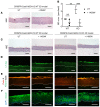
The biology of harlequin ichthyosis (HI), a devastating skin disorder caused by loss-of-function mutations in the gene ABCA12, is poorly understood, and to date, no satisfactory treatment has been developed. We sought to investigate pathomechanisms of HI that could lead to the identification of new treatments for improving patients' quality of life. In this study, RNA-Seq and functional assays were performed to define the effects of loss of ABCA12 using HI patient skin samples and an engineered CRISPR/Cas9 ABCA12 KO cell line. The HI living skin equivalent (3D model) recapitulated the HI skin phenotype. The cytokines IL-36α and IL-36γ were upregulated in HI skin, whereas the innate immune inhibitor IL-37 was strongly downregulated. We also identified STAT1 and its downstream target inducible nitric oxide synthase (NOS2) as being upregulated in the in vitro HI 3D model and HI patient skin samples. Inhibition of NOS2 using the inhibitor 1400W or the JAK inhibitor tofacitinib dramatically improved the in vitro HI phenotype by restoring the lipid barrier in the HI 3D model. Our study has identified dysregulated pathways in HI skin that are feasible therapeutic targets.
Keywords: Dermatology; Genetic diseases; Genetics; Nitric oxide; Skin.
Conflict of interest statement
Figures





Similar articles
-
Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer.J Clin Invest. 2005 Jul;115(7):1777-84. doi: 10.1172/JCI24834. J Clin Invest. 2005. PMID: 16007253 Free PMC article.
-
Harlequin ichthyosis: ABCA12 mutations underlie defective lipid transport, reduced protease regulation and skin-barrier dysfunction.Cell Tissue Res. 2013 Feb;351(2):281-8. doi: 10.1007/s00441-012-1474-9. Epub 2012 Aug 4. Cell Tissue Res. 2013. PMID: 22864982 Review.
-
Harlequin ichthyosis model mouse reveals alveolar collapse and severe fetal skin barrier defects.Hum Mol Genet. 2008 Oct 1;17(19):3075-83. doi: 10.1093/hmg/ddn204. Epub 2008 Jul 15. Hum Mol Genet. 2008. PMID: 18632686
-
A harlequin ichthyosis pig model with a novel ABCA12 mutation can be rescued by acitretin treatment.J Mol Cell Biol. 2019 Dec 19;11(12):1029-1041. doi: 10.1093/jmcb/mjz021. J Mol Cell Biol. 2019. PMID: 30925591 Free PMC article.
-
ABCA12 mutations and autosomal recessive congenital ichthyosis: a review of genotype/phenotype correlations and of pathogenetic concepts.Hum Mutat. 2010 Oct;31(10):1090-6. doi: 10.1002/humu.21326. Hum Mutat. 2010. PMID: 20672373 Review.
Cited by
-
Management of Harlequin Ichthyosis: A Brief Review of the Recent Literature.Children (Basel). 2022 Jun 15;9(6):893. doi: 10.3390/children9060893. Children (Basel). 2022. PMID: 35740830 Free PMC article. Review.
-
Efficacy and safety of secukinumab for the treatment of severe ABCA12 deficiency-related ichthyosis in a child.Skin Health Dis. 2021 May 3;1(2):e25. doi: 10.1002/ski2.25. eCollection 2021 Jun. Skin Health Dis. 2021. PMID: 35664977 Free PMC article.
-
Congenital Ichthyosis: A Practical Clinical Guide on Current Treatments and Future Perspectives.Clin Cosmet Investig Dermatol. 2023 Sep 11;16:2473-2479. doi: 10.2147/CCID.S388608. eCollection 2023. Clin Cosmet Investig Dermatol. 2023. PMID: 37719935 Free PMC article. Review.
-
Advancing novel therapies for ichthyoses.Br J Dermatol. 2021 Jun;184(6):998-999. doi: 10.1111/bjd.19698. Epub 2020 Dec 30. Br J Dermatol. 2021. PMID: 33378090 Free PMC article.
-
Design of an Integrated Microvascularized Human Skin-on-a-Chip Tissue Equivalent Model.Front Bioeng Biotechnol. 2022 Jul 19;10:915702. doi: 10.3389/fbioe.2022.915702. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35928950 Free PMC article.
References
-
- Oji V, et al. Revised nomenclature and classification of inherited ichthyoses: results of the First Ichthyosis Consensus Conference in Sorèze 2009. J Am Acad Dermatol. 2010;63(4):607–641. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

