yippee like 3 (ypel3) is a novel gene required for myelinating and perineurial glia development
- PMID: 32544203
- PMCID: PMC7319359
- DOI: 10.1371/journal.pgen.1008841
yippee like 3 (ypel3) is a novel gene required for myelinating and perineurial glia development
Erratum in
-
Correction: yippee like 3 (ypel3) is a novel gene required for myelinating and perineurial glia development.PLoS Genet. 2020 Oct 26;16(10):e1009156. doi: 10.1371/journal.pgen.1009156. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33104717 Free PMC article.
Abstract
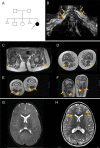
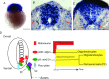
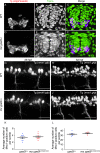
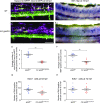
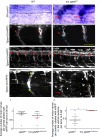
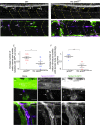
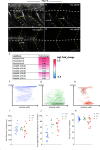
Hypomyelination, a neurological condition characterized by decreased production of myelin sheets by glial cells, often has no known etiology. Elucidating the genetic causes of hypomyelination provides a better understanding of myelination, as well as means to diagnose, council, and treat patients. Here, we present evidence that YIPPEE LIKE 3 (YPEL3), a gene whose developmental role was previously unknown, is required for central and peripheral glial cell development. We identified a child with a constellation of clinical features including cerebral hypomyelination, abnormal peripheral nerve conduction, hypotonia, areflexia, and hypertrophic peripheral nerves. Exome and genome sequencing revealed a de novo mutation that creates a frameshift in the open reading frame of YPEL3, leading to an early stop codon. We used zebrafish as a model system to validate that YPEL3 mutations are causative of neuropathy. We found that ypel3 is expressed in the zebrafish central and peripheral nervous system. Using CRISPR/Cas9 technology, we created zebrafish mutants carrying a genomic lesion similar to that of the patient. Our analysis revealed that Ypel3 is required for development of oligodendrocyte precursor cells, timely exit of the perineurial glial precursors from the central nervous system (CNS), formation of the perineurium, and Schwann cell maturation. Consistent with these observations, zebrafish ypel3 mutants have metabolomic signatures characteristic of oligodendrocyte and Schwann cell differentiation defects, show decreased levels of Myelin basic protein in the central and peripheral nervous system, and develop defasciculated peripheral nerves. Locomotion defects were observed in adult zebrafish ypel3 mutants. These studies demonstrate that Ypel3 is a novel gene required for perineurial cell development and glial myelination.
Conflict of interest statement
"The authors have declared that no competing interests exist."
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

