Disease-associated DNA2 nuclease-helicase protects cells from lethal chromosome under-replication
- PMID: 32544229
- PMCID: PMC7367196
- DOI: 10.1093/nar/gkaa524
Disease-associated DNA2 nuclease-helicase protects cells from lethal chromosome under-replication
Abstract
DNA2 is an essential nuclease-helicase implicated in DNA repair, lagging-strand DNA synthesis, and the recovery of stalled DNA replication forks (RFs). In Saccharomyces cerevisiae, dna2Δ inviability is reversed by deletion of the conserved helicase PIF1 and/or DNA damage checkpoint-mediator RAD9. It has been suggested that Pif1 drives the formation of long 5'-flaps during Okazaki fragment maturation, and that the essential function of Dna2 is to remove these intermediates. In the absence of Dna2, 5'-flaps are thought to accumulate on the lagging strand, resulting in DNA damage-checkpoint arrest and cell death. In line with Dna2's role in RF recovery, we find that the loss of Dna2 results in severe chromosome under-replication downstream of endogenous and exogenous RF-stalling. Importantly, unfaithful chromosome replication in Dna2-mutant cells is exacerbated by Pif1, which triggers the DNA damage checkpoint along a pathway involving Pif1's ability to promote homologous recombination-coupled replication. We propose that Dna2 fulfils its essential function by promoting RF recovery, facilitating replication completion while suppressing excessive RF restart by recombination-dependent replication (RDR) and checkpoint activation. The critical nature of Dna2's role in controlling the fate of stalled RFs provides a framework to rationalize the involvement of DNA2 in Seckel syndrome and cancer.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
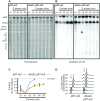





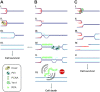
Similar articles
-
Limiting homologous recombination at stalled replication forks is essential for cell viability: DNA2 to the rescue.Curr Genet. 2020 Dec;66(6):1085-1092. doi: 10.1007/s00294-020-01106-7. Epub 2020 Sep 9. Curr Genet. 2020. PMID: 32909097 Free PMC article. Review.
-
Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta.Mol Cell Biol. 2006 Apr;26(7):2490-500. doi: 10.1128/MCB.26.7.2490-2500.2006. Mol Cell Biol. 2006. PMID: 16537895 Free PMC article.
-
Dna2 processes behind the fork long ssDNA flaps generated by Pif1 and replication-dependent strand displacement.Nat Commun. 2018 Nov 16;9(1):4830. doi: 10.1038/s41467-018-07378-5. Nat Commun. 2018. PMID: 30446656 Free PMC article.
-
Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint.Cell Cycle. 2011 May 15;10(10):1690-8. doi: 10.4161/cc.10.10.15643. Epub 2011 May 15. Cell Cycle. 2011. PMID: 21508669 Free PMC article.
-
Yet another job for Dna2: Checkpoint activation.DNA Repair (Amst). 2015 Aug;32:17-23. doi: 10.1016/j.dnarep.2015.04.009. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956863 Free PMC article. Review.
Cited by
-
Dynamic regulation of Pif1 acetylation is crucial to the maintenance of genome stability.Curr Genet. 2021 Feb;67(1):85-92. doi: 10.1007/s00294-020-01116-5. Epub 2020 Oct 20. Curr Genet. 2021. PMID: 33079209 Free PMC article. Review.
-
Canonical and novel non-canonical activities of the Holliday junction resolvase Yen1.Nucleic Acids Res. 2022 Jan 11;50(1):259-280. doi: 10.1093/nar/gkab1225. Nucleic Acids Res. 2022. PMID: 34928393 Free PMC article.
-
DNA2 knockout aggravates cerebral ischemia/reperfusion injury by reducing postsynaptic Homer1a.Zool Res. 2025 Jan 18;46(1):87-102. doi: 10.24272/j.issn.2095-8137.2024.269. Zool Res. 2025. PMID: 39846189 Free PMC article.
-
DNA2 in Chromosome Stability and Cell Survival-Is It All about Replication Forks?Int J Mol Sci. 2021 Apr 13;22(8):3984. doi: 10.3390/ijms22083984. Int J Mol Sci. 2021. PMID: 33924313 Free PMC article. Review.
-
SUMO-mediated recruitment allows timely function of the Yen1 nuclease in mitotic cells.PLoS Genet. 2022 Mar 25;18(3):e1009860. doi: 10.1371/journal.pgen.1009860. eCollection 2022 Mar. PLoS Genet. 2022. PMID: 35333860 Free PMC article.
References
-
- Bae S.H., Choi E., Lee K.H., Park J.S., Lee S.H., Seo Y.S.. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998; 273:26880–26890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

