BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment
- PMID: 32544459
- PMCID: PMC7295478
- DOI: 10.1016/j.chom.2020.05.014
BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment
Abstract
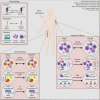
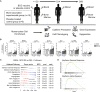
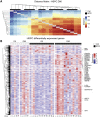
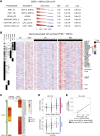
Induction of trained immunity by Bacille-Calmette-Guérin (BCG) vaccination mediates beneficial heterologous effects, but the mechanisms underlying its persistence and magnitude remain elusive. In this study, we show that BCG vaccination in healthy human volunteers induces a persistent transcriptional program connected to myeloid cell development and function within the hematopoietic stem and progenitor cell (HSPC) compartment in the bone marrow. We identify hepatic nuclear factor (HNF) family members 1a and b as crucial regulators of this transcriptional shift. These findings are corroborated by higher granulocyte numbers in BCG-vaccinated infants, HNF1 SNP variants that correlate with trained immunity, and elevated serum concentrations of the HNF1 target alpha-1 antitrypsin. Additionally, transcriptomic HSPC remodeling was epigenetically conveyed to peripheral CD14+ monocytes, displaying an activated transcriptional signature three months after BCG vaccination. Taken together, transcriptomic, epigenomic, and functional reprogramming of HSPCs and peripheral monocytes is a hallmark of BCG-induced trained immunity in humans.
Trial registration: ClinicalTrials.gov NCT01906853.
Keywords: BCG; epigenetic imprinting; hematopoietic stem cells; monocyte; myeloid cell; myelopoiesis; trained immunity; vaccination.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests M.G.N. and L.A.B.J. are scientific founders of Trained Therapeutics Discovery. All other authors declare no competing interests.
Figures



References
-
- Aaby P., Roth A., Ravn H., Napirna B.M., Rodrigues A., Lisse I.M., Stensballe L., Diness B.R., Lausch K.R., Lund N. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? J. Infect. Dis. 2011;204:245–252. - PubMed
-
- Armendariz A.D., Krauss R.M. Hepatic nuclear factor 1-alpha: inflammation, genetics, and atherosclerosis. Curr. Opin. Lipidol. 2009;20:106–111. - PubMed
-
- Arts R.J.W., Moorlag S.J.C.F.M., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100.e5. - PubMed
-
- Baldwin A.S. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

