Epigenetic Modulation of Chromatin States and Gene Expression by G-Quadruplex Structures
- PMID: 32545267
- PMCID: PMC7312119
- DOI: 10.3390/ijms21114172
Epigenetic Modulation of Chromatin States and Gene Expression by G-Quadruplex Structures
Abstract
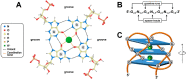
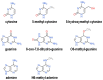
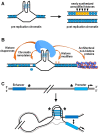
G-quadruplexes are four-stranded helical nucleic acid structures formed by guanine-rich sequences. A considerable number of studies have revealed that these noncanonical structural motifs are widespread throughout the genome and transcriptome of numerous organisms, including humans. In particular, G-quadruplexes occupy strategic locations in genomic DNA and both coding and noncoding RNA molecules, being involved in many essential cellular and organismal functions. In this review, we first outline the fundamental structural features of G-quadruplexes and then focus on the concept that these DNA and RNA structures convey a distinctive layer of epigenetic information that is critical for the complex regulation, either positive or negative, of biological activities in different contexts. In this framework, we summarize and discuss the proposed mechanisms underlying the functions of G-quadruplexes and their interacting factors. Furthermore, we give special emphasis to the interplay between G-quadruplex formation/disruption and other epigenetic marks, including biochemical modifications of DNA bases and histones, nucleosome positioning, and three-dimensional organization of chromatin. Finally, epigenetic roles of RNA G-quadruplexes in post-transcriptional regulation of gene expression are also discussed. Undoubtedly, the issues addressed in this review take on particular importance in the field of comparative epigenetics, as well as in translational research.
Keywords: DNA bases modifications; G-quadruplex; G-quartet; chromatin architecture; epigenetics; histone post-translational modifications; histone-modifying activities; noncoding RNA; nucleosome remodeling; post-transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study or the writing of the manuscript.
Figures

References
-
- Levene P.A., Jacobs W.A. Über Guanylsäure. Ber. Dtsch. Chem. Ges. 1909;42:2469–2473. doi: 10.1002/cber.190904202147. - DOI
-
- Bang I. Untersuchungen über die Guanylsäure. Biochem. Z. 1910;26:293–311.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

