Year‑Round Influenza A Virus Surveillance in Mallards (Anas platyrhynchos) Reveals Genetic Persistence During the Under‑Sampled Spring Season
- PMID: 32545281
- PMCID: PMC7354581
- DOI: 10.3390/v12060632
Year‑Round Influenza A Virus Surveillance in Mallards (Anas platyrhynchos) Reveals Genetic Persistence During the Under‑Sampled Spring Season
Abstract
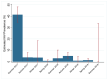
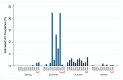
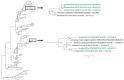
Active influenza A virus (IAV) surveillance in wild waterfowl in the United States has revolved around convenience-based sampling methods, resulting in gaps in surveillance during the spring season. We conducted active IAV surveillance in mallards continuously from July 2017 to July 2019 in the coastal marshes of Lake Erie near Port Clinton, Ohio. We aimed to understand ecological and evolutionary dynamics of IAV across multiple seasons, including the under‑sampled spring season. We collected 2096 cloacal swabs and estimated a 6.1% (95% confidence interval (CI): 0.050-0.071) prevalence during the study period. Prevalence was lowest during spring (1.0%, 95% CI: 0.004-0.015). Time‑stamped phylogenetic analyses revealed local persistence of genetic lineages of multiple gene segments. The PA segment consists of a lineage detected in multiple seasons with a time to most recent common ancestor of 2.48 years (95% highest posterior density: 2.16-2.74). Analysis of the H3 and H6 segments showed close relation between IAVs detected in spring and the following autumn migration. Though the mechanisms behind viral persistence in a single location are not well understood, we provide evidence that viruses can persist across several seasons. Current surveillance methods should be evaluated to ensure they are capturing the breadth of genetic diversity of IAV in waterfowl and prepare for IAV outbreaks in both animals and humans.
Keywords: Anas platyrhynchos; influenza A virus; mallards; phylogenetics; spring migration; surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Garten R.J., Davis C.T., Russell C.A., Shu B., Lindstrom S., Balish A., Sessions W.M., Xu X., Skepner E., Deyde V., et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

