Comparison of Chloroplast Genomes among Species of Unisexual and Bisexual Clades of the Monocot Family Araceae
- PMID: 32545339
- PMCID: PMC7355861
- DOI: 10.3390/plants9060737
Comparison of Chloroplast Genomes among Species of Unisexual and Bisexual Clades of the Monocot Family Araceae
Abstract
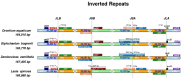
The chloroplast genome provides insight into the evolution of plant species. We de novo assembled and annotated chloroplast genomes of four genera representing three subfamilies of Araceae: Lasia spinosa (Lasioideae), Stylochaeton bogneri, Zamioculcas zamiifolia (Zamioculcadoideae), and Orontium aquaticum (Orontioideae), and performed comparative genomics using these chloroplast genomes. The sizes of the chloroplast genomes ranged from 163,770 bp to 169,982 bp. These genomes comprise 113 unique genes, including 79 protein-coding, 4 rRNA, and 30 tRNA genes. Among these genes, 17-18 genes are duplicated in the inverted repeat (IR) regions, comprising 6-7 protein-coding (including trans-splicing gene rps12), 4 rRNA, and 7 tRNA genes. The total number of genes ranged between 130 and 131. The infA gene was found to be a pseudogene in all four genomes reported here. These genomes exhibited high similarities in codon usage, amino acid frequency, RNA editing sites, and microsatellites. The oligonucleotide repeats and junctions JSB (IRb/SSC) and JSA (SSC/IRa) were highly variable among the genomes. The patterns of IR contraction and expansion were shown to be homoplasious, and therefore unsuitable for phylogenetic analyses. Signatures of positive selection were seen in three genes in S. bogneri, including ycf2, clpP, and rpl36. This study is a valuable addition to the evolutionary history of chloroplast genome structure in Araceae.
Keywords: Araceae; chloroplast genome; contraction and expansion; gene evolution; inverted repeats; phylogenetics; substitutions.
Conflict of interest statement
No conflicts of interest exists.
Figures





References
-
- Abdullah, Waseem S., Mirza B., Ahmed I., Waheed M.T. Comparative analyses of chloroplast genome of Theobroma cacao and Theobroma grandiflorum. Biologia. 2020;75:761–771. doi: 10.2478/s11756-019-00388-8. - DOI
-
- Hirao T., Watanabe A., Kurita M., Kondo T., Takata K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: Diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

