RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma
- PMID: 32545414
- PMCID: PMC7312628
- DOI: 10.3390/ijms21114190
RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma
Abstract
Pancreatic ductal adenocarcinomas (PDAC) belong to the most frequent and most deadly malignancies in the western world. Mutations in KRAS and TP53 along with some other frequent polymorphisms occur almost universally and are likely to be responsible for tumor initiation. However, these mutations cannot explain the heterogeneity in therapeutic responses observed in PDAC patients, which limits efficiency of current therapeutic strategies. Instead, recent classifications of PDAC tumor samples are based on transcriptomics data and thus include information about epigenetic, transcriptomic, and post-transcriptomic deregulations. RNA binding proteins (RBPs) are important post-transcriptional regulators involved in every aspect of the RNA life cycle and thus considerably influence the transcriptome. In this study, we systematically investigated deregulated expression, prognostic value, and essentiality reported for RBPs in PDAC or PDAC cancer models using publicly available data. We identified 44 RBPs with suggested oncogenic potential. These include various proteins, e.g., IGF2 mRNA binding proteins (IGF2BPs), with reported tumor-promoting roles. We further characterized these RBPs and found common patterns regarding their expression, interaction, and regulation by microRNAs. These analyses suggest four prime candidate oncogenic RBPs with partially validated target potential: APOBEC1, IGF2BP1 and 3, and OASL.
Keywords: APOBEC1; IGF2BP1; IGF2BP3; OASL; RNA binding proteins (RBPs); pancreatic ductal adenocarcinoma (PDAC).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

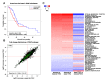


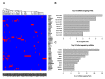

References
-
- Chen C.C., Chen L.L., Li C.P., Hsu Y.T., Jiang S.S., Fan C.S., Chua K.V., Huang S.X., Shyr Y.M., Chen L.T., et al. Myeloid-derived macrophages and secreted HSP90α induce pancreatic ductal adenocarcinoma development. Oncoimmunology. 2018;7:e1424612. doi: 10.1080/2162402X.2018.1424612. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

