Computer-Aided Ligand Discovery for Estrogen Receptor Alpha
- PMID: 32545494
- PMCID: PMC7352601
- DOI: 10.3390/ijms21124193
Computer-Aided Ligand Discovery for Estrogen Receptor Alpha
Abstract
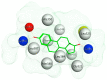
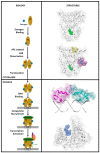
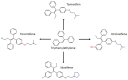
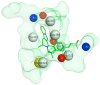
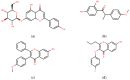
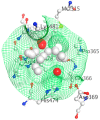
Breast cancer (BCa) is one of the most predominantly diagnosed cancers in women. Notably, 70% of BCa diagnoses are Estrogen Receptor α positive (ERα+) making it a critical therapeutic target. With that, the two subtypes of ER, ERα and ERβ, have contrasting effects on BCa cells. While ERα promotes cancerous activities, ERβ isoform exhibits inhibitory effects on the same. ER-directed small molecule drug discovery for BCa has provided the FDA approved drugs tamoxifen, toremifene, raloxifene and fulvestrant that all bind to the estrogen binding site of the receptor. These ER-directed inhibitors are non-selective in nature and may eventually induce resistance in BCa cells as well as increase the risk of endometrial cancer development. Thus, there is an urgent need to develop novel drugs with alternative ERα targeting mechanisms that can overcome the limitations of conventional anti-ERα therapies. Several functional sites on ERα, such as Activation Function-2 (AF2), DNA binding domain (DBD), and F-domain, have been recently considered as potential targets in the context of drug research and discovery. In this review, we summarize methods of computer-aided drug design (CADD) that have been employed to analyze and explore potential targetable sites on ERα, discuss recent advancement of ERα inhibitor development, and highlight the potential opportunities and challenges of future ERα-directed drug discovery.
Keywords: breast cancer; computer-aided drug design; estrogen receptor; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

