Potential Antiviral Options against SARS-CoV-2 Infection
- PMID: 32545799
- PMCID: PMC7354438
- DOI: 10.3390/v12060642
Potential Antiviral Options against SARS-CoV-2 Infection
Abstract
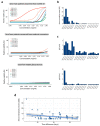
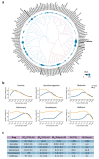
As of June 2020, the number of people infected with severe acute respiratory coronavirus 2 (SARS-CoV-2) continues to skyrocket, with more than 6.7 million cases worldwide. Both the World Health Organization (WHO) and United Nations (UN) has highlighted the need for better control of SARS-CoV-2 infections. However, developing novel virus-specific vaccines, monoclonal antibodies and antiviral drugs against SARS-CoV-2 can be time-consuming and costly. Convalescent sera and safe-in-man broad-spectrum antivirals (BSAAs) are readily available treatment options. Here, we developed a neutralization assay using SARS-CoV-2 strain and Vero-E6 cells. We identified the most potent sera from recovered patients for the treatment of SARS-CoV-2-infected patients. We also screened 136 safe-in-man broad-spectrum antivirals against the SARS-CoV-2 infection in Vero-E6 cells and identified nelfinavir, salinomycin, amodiaquine, obatoclax, emetine and homoharringtonine. We found that a combination of orally available virus-directed nelfinavir and host-directed amodiaquine exhibited the highest synergy. Finally, we developed a website to disseminate the knowledge on available and emerging treatments of COVID-19.
Keywords: antiviral drug combinations; antivirals; broad-spectrum antivirals.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- WHO WHO Publishes List of Top Emerging Diseases Likely to Cause Major Epidemics. [(accessed on 12 June 2020)]; Available online: www.who.int/medicines/ebola-treatment/WHO-list-of-top-emerging-diseases/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

