Differential Urinary Proteome Analysis for Predicting Prognosis in Type 2 Diabetes Patients with and without Renal Dysfunction
- PMID: 32545899
- PMCID: PMC7352871
- DOI: 10.3390/ijms21124236
Differential Urinary Proteome Analysis for Predicting Prognosis in Type 2 Diabetes Patients with and without Renal Dysfunction
Abstract
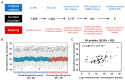
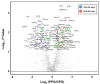
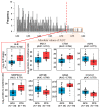
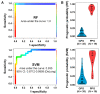
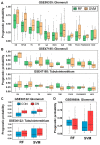
Renal dysfunction, a major complication of type 2 diabetes, can be predicted from estimated glomerular filtration rate (eGFR) and protein markers such as albumin concentration. Urinary protein biomarkers may be used to monitor or predict patient status. Urine samples were selected from patients enrolled in the retrospective diabetic kidney disease (DKD) study, including 35 with good and 19 with poor prognosis. After removal of albumin and immunoglobulin, the remaining proteins were reduced, alkylated, digested, and analyzed qualitatively and quantitatively with a nano LC-MS platform. Each protein was identified, and its concentration normalized to that of creatinine. A prognostic model of DKD was formulated based on the adjusted quantities of each protein in the two groups. Of 1296 proteins identified in the 54 urine samples, 66 were differentially abundant in the two groups (area under the curve (AUC): p-value < 0.05), but none showed significantly better performance than albumin. To improve the predictive power by multivariate analysis, five proteins (ACP2, CTSA, GM2A, MUC1, and SPARCL1) were selected as significant by an AUC-based random forest method. The application of two classifiers-support vector machine and random forest-showed that the multivariate model performed better than univariate analysis of mucin-1 (AUC: 0.935 vs. 0.791) and albumin (AUC: 1.0 vs. 0.722). The urinary proteome can reflect kidney function directly and can predict the prognosis of patients with chronic kidney dysfunction. Classification based on five urinary proteins may better predict the prognosis of DKD patients than urinary albumin concentration or eGFR.
Keywords: diabetic kidney disease; kidney function; machine learning; mass spectrometry; proteomics; statistical clinical model; urine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Ahn J.H., Yu J.H., Ko S.H., Kwon H.S., Kim D.J., Kim J.H., Kim C.S., Song K.H., Won J.C., Lim S., et al. Prevalence and determinants of diabetic nephropathy in Korea: Korea national health and nutrition examination survey. Diabetes Metab. J. 2014;38:109–119. doi: 10.4093/dmj.2014.38.2.109. - DOI - PMC - PubMed
-
- Tuttle K.R., Bakris G.L., Bilous R.W., Chiang J.L., de Boer I.H., Goldstein-Fuchs J., Hirsch I.B., Kalantar-Zadeh K., Narva A.S., Navaneethan S.D., et al. Diabetic kidney disease: A report from an ADA Consensus Conference. Diabetes Care. 2014;37:2864–2883. doi: 10.2337/dc14-1296. - DOI - PMC - PubMed
-
- Collins A.J., Foley R.N., Chavers B., Gilbertson D., Herzog C., Johansen K., Kasiske B., Kutner N., Liu J., St Peter W., et al. United States Renal Data System 2011 Annual Data Report: Atlas of chronic kidney disease & end-stage renal disease in the United States. Am. J. Kidney Dis. 2012;59:A7. doi: 10.1053/j.ajkd.2011.11.015. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

