A high-coverage Neandertal genome from Chagyrskaya Cave
- PMID: 32546518
- PMCID: PMC7334501
- DOI: 10.1073/pnas.2004944117
A high-coverage Neandertal genome from Chagyrskaya Cave
Abstract
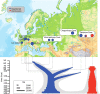
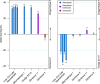
We sequenced the genome of a Neandertal from Chagyrskaya Cave in the Altai Mountains, Russia, to 27-fold genomic coverage. We show that this Neandertal was a female and that she was more related to Neandertals in western Eurasia [Prüfer et al., Science 358, 655-658 (2017); Hajdinjak et al., Nature 555, 652-656 (2018)] than to Neandertals who lived earlier in Denisova Cave [Prüfer et al., Nature 505, 43-49 (2014)], which is located about 100 km away. About 12.9% of the Chagyrskaya genome is spanned by homozygous regions that are between 2.5 and 10 centiMorgans (cM) long. This is consistent with the fact that Siberian Neandertals lived in relatively isolated populations of less than 60 individuals. In contrast, a Neandertal from Europe, a Denisovan from the Altai Mountains, and ancient modern humans seem to have lived in populations of larger sizes. The availability of three Neandertal genomes of high quality allows a view of genetic features that were unique to Neandertals and that are likely to have been at high frequency among them. We find that genes highly expressed in the striatum in the basal ganglia of the brain carry more amino-acid-changing substitutions than genes expressed elsewhere in the brain, suggesting that the striatum may have evolved unique functions in Neandertals.
Keywords: Neandertals; genetics; genome; human evolution.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

