High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors
- PMID: 32546544
- PMCID: PMC8023011
- DOI: 10.1126/scisignal.aay1451
High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors
Abstract
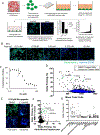
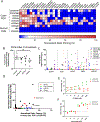
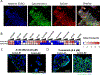
Despite decades of effort, the sensitivity of patient tumors to individual drugs is often not predictable on the basis of molecular markers alone. Therefore, unbiased, high-throughput approaches to match patient tumors to effective drugs, without requiring a priori molecular hypotheses, are critically needed. Here, we improved upon a method that we previously reported and developed called high-throughput dynamic BH3 profiling (HT-DBP). HT-DBP is a microscopy-based, single-cell resolution assay that enables chemical screens of hundreds to thousands of candidate drugs on freshly isolated tumor cells. The method identifies chemical inducers of mitochondrial apoptotic signaling, a mechanism of cell death. HT-DBP requires only 24 hours of ex vivo culture, which enables a more immediate study of fresh primary tumor cells and minimizes adaptive changes that occur with prolonged ex vivo culture. Effective compounds identified by HT-DBP induced tumor regression in genetically engineered and patient-derived xenograft (PDX) models of breast cancer. We additionally found that chemical vulnerabilities changed as cancer cells expanded ex vivo. Furthermore, using PDX models of colon cancer and resected tumors from colon cancer patients, our data demonstrated that HT-DBP could be used to generate personalized pharmacotypes. Thus, HT-DBP appears to be an ex vivo functional method with sufficient scale to simultaneously function as a companion diagnostic, therapeutic personalization, and discovery tool.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, and Meyerson M.. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. - PubMed
-
- Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, Raghavan S, Kim J, Brais LK, Ragon D, Welch MW, Reilly E, McCabe D, Marini L, Anderka K, Helvie K, Oliver N, Babic A, Da Silva A, Nadres B, Van Seventer EE, Shahzade HA, St Pierre JP, Burke KP, Clancy T, Cleary JM, Doyle LA, Jajoo K, McCleary NJ, Meyerhardt JA, Murphy JE, Ng K, Patel AK, Perez K, Rosenthal MH, Rubinson DA, Ryou M, Shapiro GI, Sicinska E, Silverman SG, Nagy RJ, Lanman RB, Knoerzer D, Welsch DJ, Yurgelun MB, Fuchs CS, Garraway LA, Getz G, Hornick JL, Johnson BE, Kulke MH, Mayer RJ, Miller JW, Shyn PB, Tuveson DA, Wagle N, Yeh JJ, Hahn WC, Corcoran RB, Carter SL, and Wolpin BM. 2018. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov 8: 1096–1111. - PMC - PubMed
-
- Letai A. 2017. Functional precision cancer medicine-moving beyond pure genomics. Nat Med 23: 1028–1035. - PubMed
-
- Burstein HJ, Mangu PB, Somerfield MR, Schrag D, Samson D, Holt L, Zelman D, Ajani JA, and American O. Society of Clinical. 2011. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29: 3328–3330. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

