Assessment of Luminal and Basal Phenotypes in Bladder Cancer
- PMID: 32546765
- PMCID: PMC7298008
- DOI: 10.1038/s41598-020-66747-7
Assessment of Luminal and Basal Phenotypes in Bladder Cancer
Abstract
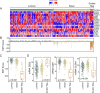
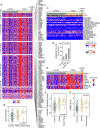
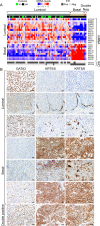
Genomic profiling studies have demonstrated that bladder cancer can be divided into two molecular subtypes referred to as luminal and basal with distinct clinical behaviors and sensitivities to frontline chemotherapy. We analyzed the mRNA expressions of signature luminal and basal genes in bladder cancer tumor samples from publicly available and MD Anderson Cancer Center cohorts. We developed a quantitative classifier referred to as basal to luminal transition (BLT) score which identified the molecular subtypes of bladder cancer with 80-94% sensitivity and 83-93% specificity. In order to facilitate molecular subtyping of bladder cancer in primary care centers, we analyzed the protein expressions of signature luminal (GATA3) and basal (KRT5/6) markers by immunohistochemistry, which identified molecular subtypes in over 80% of the cases. In conclusion, we provide a tool for assessment of molecular subtypes of bladder cancer in routine clinical practice.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

