Knockdown of circRAD18 Mitigates Breast Cancer Progression through the Regulation of miR-613/HK2 Axis
- PMID: 32547203
- PMCID: PMC7245444
- DOI: 10.2147/CMAR.S243300
Knockdown of circRAD18 Mitigates Breast Cancer Progression through the Regulation of miR-613/HK2 Axis
Retraction in
-
Knockdown of circRAD18 Mitigates Breast Cancer Progression through the Regulation of miR-613/HK2 Axis [Retraction].Cancer Manag Res. 2022 Sep 22;14:2859-2860. doi: 10.2147/CMAR.S390081. eCollection 2022. Cancer Manag Res. 2022. PMID: 36171863 Free PMC article.
Abstract
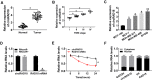
Background: Breast cancer (BC) remains the most prevalent malignancy and the leading cause of cancer death. Circular RNAs (circRNAs) have been discovered to serve as crucial regulators in BC. In the current work, we aimed to study the impact of circRAD18 (hsa_circ_0002453) on BC progression and mechanism governing it.
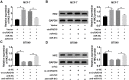
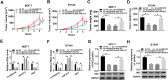
Materials and methods: The expression levels of circRAD18, miR-613 and hexokinase 2 (HK2) mRNA were determined by quantitative real-time polymerase chain reaction (qRT-PCR). CircRAD18 identification was performed using RNase R digestion and actinomycin D assay. Cell viability, colony formation, apoptosis, migration, invasion and glycolysis were measured by Cell Counting Kit-8 assay, colony formation assay, flow cytometry, transwell analysis and extracellular acidification rate detection assay, respectively. Western blot was used to assess the levels of E-Cadherin, Vimentin, N-Cadherin and HK2 protein. The targeted interplay between miR-613 and circRAD18 or HK2 was detected by dual-luciferase reporter assay. Xenograft model assay was performed to observe the role of circRAD18 in vivo.
Results: CircRAD18 was highly expressed in BC tissues and cells. CircRAD18 depletion hindered BC cell malignant behaviors, as evidenced by the inhibition in cell viability, colony formation, migration, invasion, epithelial to mesenchymal transition and glycolysis, as well as the promotion in cell apoptosis. CircRAD18 directly interacted with miR-613, and miR-613 mediated the repressive effect of circRAD18 knockdown on BC cell malignant behaviors. Moreover, HK2 was a direct target of miR-613, and circRAD18 positively regulated HK2 expression via sponging miR-613. Additionally, circRAD18 knockdown repressed tumor growth in vivo by miR-613.
Conclusion: Our current work suggested that circRAD18 silencing suppressed BC cell malignant behaviors in vitro and tumor growth in vivo at least partly via the regulation of the miR-613/HK2 axis, highlighting that circRAD18 might be a promising therapeutic target for BC treatment.
Keywords: BC; HK2; circRAD18; malignant behaviors; miR-613.
© 2020 Zang et al.
Conflict of interest statement
The authors declare that they have no financial conflicts of interest.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

