Fungal Mitogenomes: Relevant Features to Planning Plant Disease Management
- PMID: 32547508
- PMCID: PMC7272585
- DOI: 10.3389/fmicb.2020.00978
Fungal Mitogenomes: Relevant Features to Planning Plant Disease Management
Abstract
Mitochondrial genomes (mt-genomes) are characterized by a distinct codon usage and their autonomous replication. Mt-genomes encode highly conserved genes (mt-genes), like proteins involved in electron transport and oxidative phosphorylation but they also carry highly variable regions that are in part responsible for their high plasticity. The degree of conservation of their genes is such that they allow the establishment of phylogenetic relationships even across distantly related species. Here, we describe the mechanisms that generate changes along mt-genomes, which play key roles at enlarging the ability of fungi to adapt to changing environments. Within mt-genomes of fungal pathogens, there are dispensable as well as indispensable genes for survival, virulence and/or pathogenicity. We also describe the different complexes or mechanisms targeted by fungicides, thus addressing a relevant issue regarding disease management. Despite the controversial origin and evolution of fungal mt-genomes, the intrinsic mechanisms and molecular biology involved in their evolution will help to understand, at the molecular level, the strategies for fungal disease management.
Keywords: fungal interactions; fungal mitogenome; mobile elements; pathogenesis; pathogens; plant disease management; virulence.
Copyright © 2020 Medina, Franco, Bartel, Martinez Alcántara, Saparrat and Balatti.
Figures
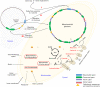
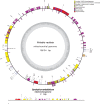
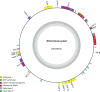
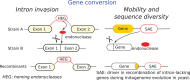
References
-
- Arcilla J. E. A., Arango R. E., Torres J. M., Arias T. (2019). Comparative genomics in plant fungal pathogens (Mycosphaerellaceae): variation in mitochondrial composition due to at least five independent intron invasions. bioRxiv. [Preprint] 10.1101/694562 - DOI
Publication types
LinkOut - more resources
Full Text Sources

